3H2Y
 
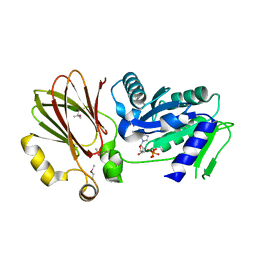 | | Crystal structure of YqeH GTPase from Bacillus anthracis with dGDP bound | | Descriptor: | 2'-DEOXYGUANOSINE-5'-DIPHOSPHATE, GTPase family protein | | Authors: | Brunzelle, J.S, Anderson, S.M, Xu, X, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-04-15 | | Release date: | 2009-06-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of YqeH GTPase from Bacillus anthracis with dGDP Bound
To be Published
|
|
3GEU
 
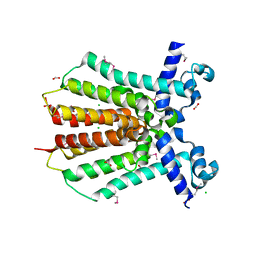 | | Crystal Structure of IcaR from Staphylococcus aureus, a member of the tetracycline repressor protein family | | Descriptor: | CHLORIDE ION, FORMIC ACID, Intercellular adhesion protein R, ... | | Authors: | Anderson, S.M, Brunzelle, J.S, Wawrzak, Z, Skarina, T, Papazisi, L, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-02-26 | | Release date: | 2009-03-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of IcaR from Staphylococcus aureus, a member of the tetracycline repressor protein family
To be Published
|
|
3FF1
 
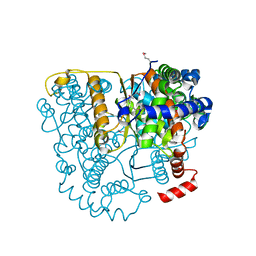 | | Structure of Glucose 6-phosphate Isomerase from Staphylococcus aureus | | Descriptor: | GLUCOSE-6-PHOSPHATE, Glucose-6-phosphate isomerase, SODIUM ION | | Authors: | Anderson, S.M, Brunzelle, J.S, Onopriyenko, O, Peterson, S, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2008-12-01 | | Release date: | 2009-02-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure of Glucose 6-phosphate Isomerase from Staphylococcus aureus
TO BE PUBLISHED
|
|
3HZN
 
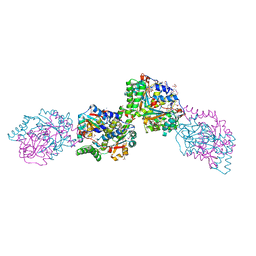 | | Structure of the Salmonella typhimurium nfnB dihydropteridine reductase | | Descriptor: | ACETATE ION, CHLORIDE ION, CITRATE ANION, ... | | Authors: | Anderson, S.M, Wawrzak, Z, Onopriyenko, O, Skarina, T, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-06-23 | | Release date: | 2009-07-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the Salmonella typhimurium nfnB dihydropteridine reductase
TO BE PUBLISHED
|
|
3IHS
 
 | | Crystal Structure of a Phosphocarrier Protein HPr from Bacillus anthracis str. Ames | | Descriptor: | Phosphocarrier protein HPr | | Authors: | Brunzelle, J.S, Anderson, S.M, Skarina, T, Onopriyenko, O, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-07-30 | | Release date: | 2009-08-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Crystal Structure of a Phosphocarrier Protein HPr from Bacillus anthracis str. Ames
To be Published
|
|
3IG4
 
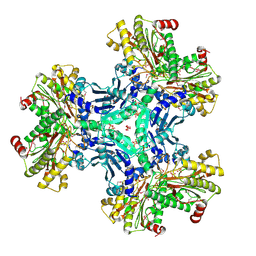 | | Structure of a putative aminopeptidase P from Bacillus anthracis | | Descriptor: | MANGANESE (II) ION, SULFATE ION, Xaa-pro aminopeptidase | | Authors: | Anderson, S.M, Wawrzak, Z, Skarina, T, Onopriyenko, O, Kwon, K, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-07-27 | | Release date: | 2009-08-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structure of a putative aminopeptidase P from Bacillus anthracis
To be Published
|
|
3HJV
 
 | | 1.7 Angstrom resolution crystal structure of an acyl carrier protein S-malonyltransferase from Vibrio cholerae O1 biovar eltor str. N16961 | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, CHLORIDE ION, Malonyl Coa-acyl carrier protein transacylase, ... | | Authors: | Halavaty, A.S, Wawrzak, Z, Anderson, S, Skarina, T, Onopriyenko, O, Kwon, K, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-05-22 | | Release date: | 2009-06-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 1.7 Angstrom resolution crystal structure of an acyl carrier protein S-malonyltransferase from Vibrio cholerae O1 biovar eltor str. N16961
To be Published
|
|
3N8H
 
 | | Crystal Structure of Pantoate-beta-alanine Ligase from Francisella tularensis | | Descriptor: | ACETIC ACID, ADENOSINE MONOPHOSPHATE, GLYCEROL, ... | | Authors: | Maltseva, N, Kim, Y, Papazisi, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-05-28 | | Release date: | 2010-06-16 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Crystal Structure of Pantoate-beta-alanine Ligase from Francisella tularensis
To be Published, 2010
|
|
4MBR
 
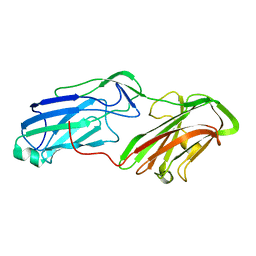 | | 3.65 Angstrom Crystal Structure of Serine-rich Repeat Protein (Srr2) from Streptococcus agalactiae | | Descriptor: | Serine-rich repeat protein 2 | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Seo, H.S, Seepersaud, R, Doran, K.S, Iverson, T.M, Sullam, P.M, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-08-19 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | Characterization of Fibrinogen Binding by Glycoproteins Srr1 and Srr2 of Streptococcus agalactiae.
J.Biol.Chem., 288, 2013
|
|
4MA0
 
 | | The crystal structure of phosphoribosylaminoimidazole carboxylase ATPase subunit of Francisella tularensis subsp. tularensis SCHU S4 in complex with partially hydrolysed ATP | | Descriptor: | ADENOSINE MONOPHOSPHATE, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Tan, K, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-08-15 | | Release date: | 2013-08-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.982 Å) | | Cite: | The crystal structure of phosphoribosylaminoimidazole carboxylase ATPase subunit of Francisella tularensis subsp. tularensis SCHU S4 in complex with partially hydrolysed ATP
To be Published
|
|
4MFG
 
 | | 2.0 Angstrom Resolution Crystal Structure of Putative Carbonic Anhydrase from Clostridium difficile. | | Descriptor: | MAGNESIUM ION, NICKEL (II) ION, Putative acyltransferase | | Authors: | Minasov, G, Wawrzak, Z, Kudritska, M, Grimshaw, S, Kwon, K, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-08-27 | | Release date: | 2013-09-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 2.0 Angstrom Resolution Crystal Structure of Putative Carbonic Anhydrase from Clostridium difficile.
TO BE PUBLISHED
|
|
4MHD
 
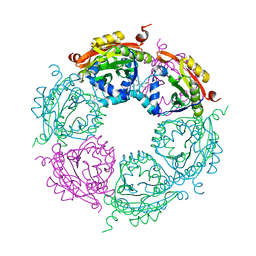 | | Crystal structure of spermidine N-acetyltransferase from Vibrio cholerae in complex with spermidine | | Descriptor: | SPERMIDINE, Spermidine n1-acetyltransferase | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Kuhn, M.L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-08-29 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | A Novel Polyamine Allosteric Site of SpeG from Vibrio cholerae Is Revealed by Its Dodecameric Structure.
J.Mol.Biol., 427, 2015
|
|
4MAM
 
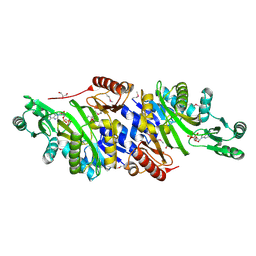 | | The crystal structure of phosphoribosylaminoimidazole carboxylase ATPase subunit of Francisella tularensis subsp. tularensis SCHU S4 in complex with an ADP analog, AMP-CP | | Descriptor: | GLYCEROL, PHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, Phosphoribosylaminoimidazole carboxylase, ... | | Authors: | Tan, K, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-08-16 | | Release date: | 2013-08-28 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.474 Å) | | Cite: | The crystal structure of phosphoribosylaminoimidazole carboxylase ATPase subunit of Francisella tularensis subsp. tularensis SCHU S4 in complex with an ADP analog, AMP-CP
To be Published
|
|
4MBO
 
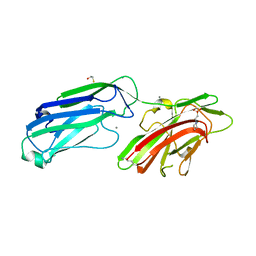 | | 1.65 Angstrom Crystal Structure of Serine-rich Repeat Adhesion Glycoprotein (Srr1) from Streptococcus agalactiae | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, BETA-MERCAPTOETHANOL, CALCIUM ION, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Seo, H.S, Seepersaud, R, Doran, K.S, Iverson, T.M, Sullam, P.M, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-08-19 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Characterization of Fibrinogen Binding by Glycoproteins Srr1 and Srr2 of Streptococcus agalactiae.
J.Biol.Chem., 288, 2013
|
|
4M8I
 
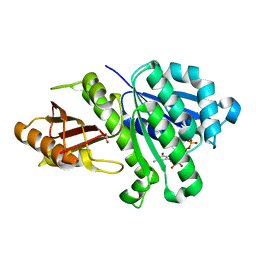 | | 1.43 Angstrom resolution crystal structure of cell division protein FtsZ (ftsZ) from Staphylococcus epidermidis RP62A in complex with GDP | | Descriptor: | Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE, SULFATE ION | | Authors: | Halavaty, A.S, Minasov, G, Winsor, J, Dubrovska, I, Filippova, E.V, Olsen, D.B, Therien, A, Shuvalova, L, Young, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-08-13 | | Release date: | 2013-09-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | 1.43 Angstrom resolution crystal structure of cell division protein FtsZ (ftsZ) from Staphylococcus epidermidis RP62A in complex with GDP
To be Published
|
|
4LRL
 
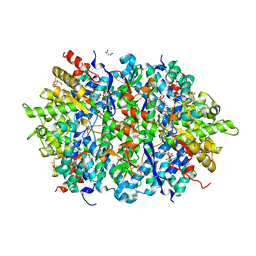 | | Structure of an Enterococcus Faecalis HD-domain protein complexed with dGTP and dTTP | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, HD domain protein, ... | | Authors: | Vorontsov, I.I, Minasov, G, Shuvalova, L, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-07-19 | | Release date: | 2013-11-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Mechanisms of Allosteric Activation and Inhibition of the Deoxyribonucleoside Triphosphate Triphosphohydrolase from Enterococcus faecalis.
J.Biol.Chem., 289, 2014
|
|
3NZT
 
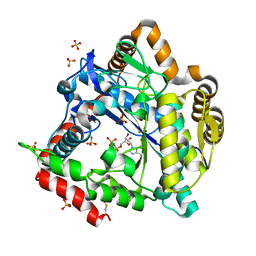 | | 2.0 Angstrom Crystal structure of Glutamate--Cysteine Ligase (gshA) ftom Francisella tularensis in Complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Glutamate--cysteine ligase, SULFATE ION | | Authors: | Minasov, G, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-07-16 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 2.0 Angstrom Crystal structure of Glutamate--Cysteine Ligase (gshA) ftom Francisella tularensis in Complex with AMP.
To be Published
|
|
3O2R
 
 | | Structural flexibility in region involved in dimer formation of nuclease domain of Ribonuclase III (rnc) from Campylobacter jejuni | | Descriptor: | CHLORIDE ION, Ribonuclease III | | Authors: | Minasov, G, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-07-22 | | Release date: | 2010-08-04 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.251 Å) | | Cite: | Structural Flexibility in Region Involved in Dimer Formation of Nuclease Domain of Ribonuclase III (rnc) from Campylobacter jejuni.
TO BE PUBLISHED
|
|
4M9U
 
 | | The crystal structure of phosphoribosylaminoimidazole carboxylase ATPase subunit of Francisella tularensis subsp. tularensis SCHU S4 | | Descriptor: | ACETATE ION, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Tan, K, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-08-15 | | Release date: | 2013-08-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | The crystal structure of phosphoribosylaminoimidazole carboxylase ATPase subunit of Francisella tularensis subsp. tularensis SCHU S4
To be Published
|
|
3O6D
 
 | | Pyridoxal phosphate biosynthetic protein PdxJ from Campylobacter jejuni in complex with pyridoxine-5'-phosphate | | Descriptor: | PHOSPHATE ION, PYRIDOXINE-5'-PHOSPHATE, Pyridoxine 5'-phosphate synthase | | Authors: | Osipiuk, J, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-07-28 | | Release date: | 2010-08-11 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | X-ray crystal structure of pyridoxal phosphate biosynthetic protein PdxJ from Campylobacter jejuni.
To be Published
|
|
3O6V
 
 | | Crystal structure of Uridine Phosphorylase from Vibrio cholerae O1 biovar El Tor | | Descriptor: | FORMIC ACID, GLYCEROL, Uridine phosphorylase | | Authors: | Maltseva, N, Kim, Y, Hasseman, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-07-29 | | Release date: | 2010-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.695 Å) | | Cite: | Crystal structure of Uridine Phosphorylase from Vibrio cholerae O1 biovar El Tor
To be Published
|
|
3NNT
 
 | | Crystal Structure of K170M Mutant of Type I 3-Dehydroquinate Dehydratase (aroD) from Salmonella typhimurium LT2 in Non-Covalent Complex with Dehydroquinate. | | Descriptor: | 1,3,4-TRIHYDROXY-5-OXO-CYCLOHEXANECARBOXYLIC ACID, 3-dehydroquinate dehydratase | | Authors: | Minasov, G, Light, S.H, Shuvalova, L, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-06-24 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Insights into the mechanism of type I dehydroquinate dehydratases from structures of reaction intermediates.
J.Biol.Chem., 286, 2011
|
|
4MGE
 
 | | 1.85 Angstrom Resolution Crystal Structure of PTS System Cellobiose-specific Transporter Subunit IIB from Bacillus anthracis. | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, PTS system, ... | | Authors: | Minasov, G, Shuvalova, L, Halavaty, A, Winsor, J, Dubrovska, I, Shatsman, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-08-28 | | Release date: | 2013-09-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | 1.85 Angstrom Resolution Crystal Structure of PTS System Cellobiose-specific Transporter Subunit IIB from Bacillus anthracis.
TO BE PUBLISHED
|
|
4MA5
 
 | | The crystal structure of phosphoribosylaminoimidazole carboxylase ATPase subunit of Francisella tularensis subsp. tularensis SCHU S4 in complex with an ATP analog, AMP-PNP. | | Descriptor: | FORMIC ACID, GLYCEROL, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Tan, K, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-08-15 | | Release date: | 2013-08-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.809 Å) | | Cite: | The crystal structure of phosphoribosylaminoimidazole carboxylase ATPase subunit of Francisella tularensis subsp. tularensis SCHU S4 in complex with an ATP analog, AMP-PNP.
To be Published
|
|
3O6C
 
 | | Pyridoxal phosphate biosynthetic protein PdxJ from Campylobacter jejuni | | Descriptor: | PHOSPHATE ION, Pyridoxine 5'-phosphate synthase | | Authors: | Osipiuk, J, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-07-28 | | Release date: | 2010-08-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Pyridoxal phosphate biosynthetic protein PdxJ from Campylobacter jejuni.
To be Published
|
|
