6ALV
 
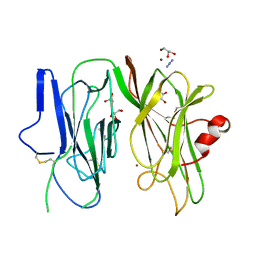 | | Crystal structure of H107A-peptidylglycine alpha-hydroxylating monooxygenase (PHM) mutant (no CuH bound) | | Descriptor: | AZIDE ION, COPPER (II) ION, GLYCEROL, ... | | Authors: | Maheshwari, S, Rudzka, K, Gabelli, S.B, Amzel, L.M. | | Deposit date: | 2017-08-08 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Effects of copper occupancy on the conformational landscape of peptidylglycine alpha-hydroxylating monooxygenase.
Commun Biol, 1, 2018
|
|
6AN3
 
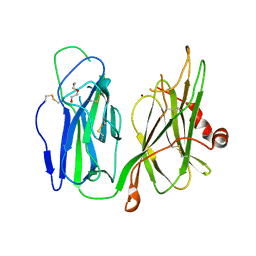 | | Crystal structure of H172A-peptidylglycine alpha-hydroxylating monooxygenase (PHM) mutant soaked with peptide (no CuH bound, no peptide bound) | | Descriptor: | COPPER (II) ION, DI(HYDROXYETHYL)ETHER, Peptidyl-glycine alpha-amidating monooxygenase | | Authors: | Maheshwari, S, Rudzka, K, Gabelli, S.B, Amzel, L.M. | | Deposit date: | 2017-08-11 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Effects of copper occupancy on the conformational landscape of peptidylglycine alpha-hydroxylating monooxygenase.
Commun Biol, 1, 2018
|
|
6AY0
 
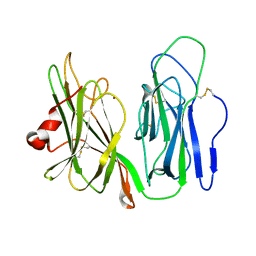 | | Crystal structure of H108A peptidylglycine alpha-hydroxylating monooxygenase (PHM) soaked with peptide | | Descriptor: | COPPER (II) ION, Peptidyl-glycine alpha-amidating monooxygenase | | Authors: | Maheshwari, S, Rudzka, K, Gabelli, S.B, Amzel, L.M. | | Deposit date: | 2017-09-07 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Effects of copper occupancy on the conformational landscape of peptidylglycine alpha-hydroxylating monooxygenase.
Commun Biol, 1, 2018
|
|
6AMP
 
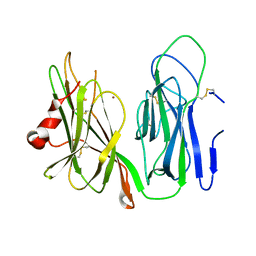 | | Crystal structure of H172A PHM (CuH absent, CuM present) | | Descriptor: | COPPER (II) ION, Peptidyl-glycine alpha-amidating monooxygenase | | Authors: | Maheshwari, S, Rudzka, K, Gabelli, S.B, Amzel, L.M. | | Deposit date: | 2017-08-10 | | Release date: | 2018-07-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Effects of copper occupancy on the conformational landscape of peptidylglycine alpha-hydroxylating monooxygenase.
Commun Biol, 1, 2018
|
|
6AO6
 
 | | Crystal structure of H108A peptidylglycine alpha-hydroxylating monooxygenase (PHM) | | Descriptor: | COPPER (II) ION, GLYCEROL, NICKEL (II) ION, ... | | Authors: | Maheshwari, S, Rudzka, K, Gabelli, S.B, Amzel, L.M. | | Deposit date: | 2017-08-15 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Effects of copper occupancy on the conformational landscape of peptidylglycine alpha-hydroxylating monooxygenase.
Commun Biol, 1, 2018
|
|
6E0T
 
 | | C-terminal domain of Fission Yeast OFD1 | | Descriptor: | Prolyl 3,4-dihydroxylase ofd1 | | Authors: | Bianchet, M.A, Amzel, L.M, Espenshade, P.J, Yeh, T. | | Deposit date: | 2018-07-06 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | The hypoxic regulator of sterol synthesis nro1 is a nuclear import adaptor.
Structure, 19, 2011
|
|
1OPM
 
 | |
1PHM
 
 | |
1Q3F
 
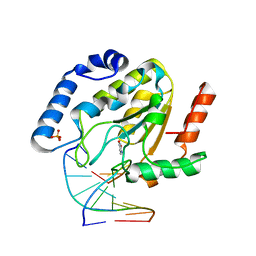 | | Uracil DNA glycosylase bound to a cationic 1-aza-2'-deoxyribose-containing DNA | | Descriptor: | 5'-D(*AP*AP*AP*GP*AP*TP*AP*AP*CP*A)-3', 5'-D(*TP*GP*TP*(NRI)P*AP*TP*CP*TP*T)-3', PHOSPHATE ION, ... | | Authors: | Bianchet, M.A, Seiple, L.A, Jiang, Y.L, Ichikawa, Y, Amzel, L.M, Stivers, J.T. | | Deposit date: | 2003-07-29 | | Release date: | 2004-03-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Electrostatic guidance of glycosyl cation migration along the reaction coordinate of uracil DNA glycosylase.
Biochemistry, 42, 2003
|
|
1QR2
 
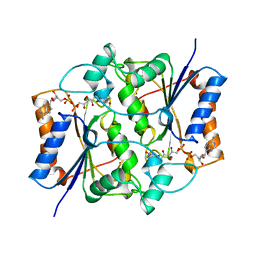 | | HUMAN QUINONE REDUCTASE TYPE 2 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, PROTEIN (QUINONE REDUCTASE TYPE 2), ZINC ION | | Authors: | Foster, C, Bianchet, M.A, Talalay, P, Amzel, L.M. | | Deposit date: | 1999-04-15 | | Release date: | 1999-08-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of human quinone reductase type 2, a metalloflavoprotein.
Biochemistry, 38, 1999
|
|
1QRD
 
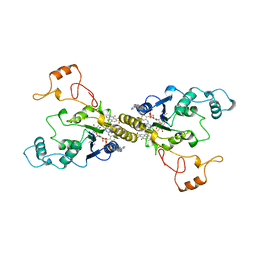 | | QUINONE REDUCTASE/FAD/CIBACRON BLUE/DUROQUINONE COMPLEX | | Descriptor: | CIBACRON BLUE, DUROQUINONE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Li, R, Bianchet, M.A, Talalay, P, Amzel, L.M. | | Deposit date: | 1995-07-28 | | Release date: | 1996-10-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The three-dimensional structure of NAD(P)H:quinone reductase, a flavoprotein involved in cancer chemoprotection and chemotherapy: mechanism of the two-electron reduction.
Proc.Natl.Acad.Sci.USA, 92, 1995
|
|
1RYA
 
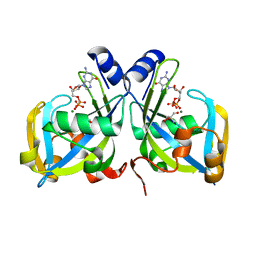 | | Crystal Structure of the E. coli GDP-mannose mannosyl hydrolase in complex with GDP and MG | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, GDP-mannose mannosyl hydrolase, ... | | Authors: | Gabelli, S.B, Bianchet, M.A, Legler, P.M, Mildvan, A.S, Amzel, L.M. | | Deposit date: | 2003-12-20 | | Release date: | 2004-06-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure and mechanism of GDP-mannose glycosyl hydrolase, a Nudix enzyme that cleaves at carbon instead of phosphorus.
Structure, 12, 2004
|
|
1SDW
 
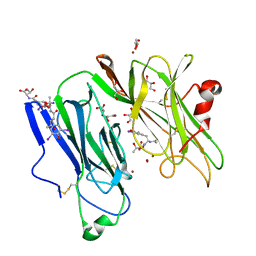 | | Reduced (Cu+) peptidylglycine alpha-hydroxylating monooxygenase with bound peptide and dioxygen | | Descriptor: | COPPER (II) ION, GLYCEROL, N-ALPHA-ACETYL-3,5-DIIODOTYROSYL-D-THREONINE, ... | | Authors: | Prigge, S.T, Eipper, B.A, Mains, R.E, Amzel, L.M. | | Deposit date: | 2004-02-14 | | Release date: | 2004-05-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Dioxygen binds end-on to mononuclear copper in a precatalytic enzyme complex.
Science, 304, 2004
|
|
3CQO
 
 | |
3CK0
 
 | |
3EEU
 
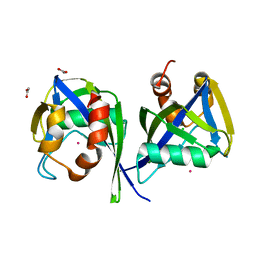 | | Structure of the RNA pyrophosphohydrolase BdRppH in complex with Holmium | | Descriptor: | ACETATE ION, CHLORIDE ION, HOLMIUM ATOM, ... | | Authors: | Messing, S.A, Gabelli, S.B, Amzel, L.M. | | Deposit date: | 2008-09-05 | | Release date: | 2009-03-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and Biological Function of the RNA Pyrophosphohydrolase BdRppH from Bdellovibrio bacteriovorus.
Structure, 17, 2009
|
|
3EES
 
 | |
3EF5
 
 | |
3FFU
 
 | |
3FW0
 
 | | Structure of Peptidyl-alpha-hydroxyglycine alpha-Amidating Lyase (PAL) bound to alpha-hydroxyhippuric acid (non-peptidic substrate) | | Descriptor: | (2S)-hydroxy[(phenylcarbonyl)amino]ethanoic acid, CALCIUM ION, MERCURY (II) ION, ... | | Authors: | Chufan, E.E, De, M, Eipper, B.A, Mains, R.E, Amzel, L.M. | | Deposit date: | 2009-01-16 | | Release date: | 2009-09-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Amidation of bioactive peptides: the structure of the lyase domain of the amidating enzyme.
Structure, 17, 2009
|
|
3FVZ
 
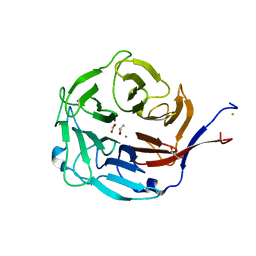 | | Structure of Peptidyl-alpha-hydroxyglycine alpha-Amidating Lyase (PAL) | | Descriptor: | ACETATE ION, CALCIUM ION, FE (III) ION, ... | | Authors: | Chufan, E.E, De, M, Eipper, B.A, Mains, R.E, Amzel, L.M. | | Deposit date: | 2009-01-16 | | Release date: | 2009-09-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Amidation of bioactive peptides: the structure of the lyase domain of the amidating enzyme.
Structure, 17, 2009
|
|
3FZD
 
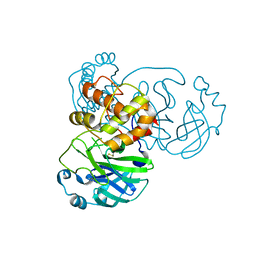 | | Mutation of Asn28 disrupts the enzymatic activity and dimerization of SARS 3CLpro | | Descriptor: | 3C-like proteinase | | Authors: | Barrila, J, Gabelli, S, Bacha, U, Amzel, L.M, Freire, E. | | Deposit date: | 2009-01-25 | | Release date: | 2010-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Mutation of Asn28 disrupts the dimerization and enzymatic activity of SARS 3CL(pro) .
Biochemistry, 49, 2010
|
|
1K12
 
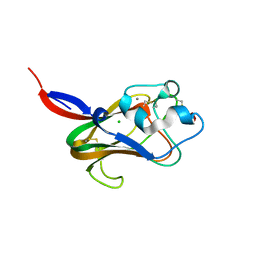 | | Fucose Binding lectin | | Descriptor: | CALCIUM ION, CHLORIDE ION, LECTIN, ... | | Authors: | Bianchet, M.A, Odom, E.W, Vasta, G.R, Amzel, L.M. | | Deposit date: | 2001-09-23 | | Release date: | 2002-07-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A novel fucose recognition fold involved in innate immunity.
Nat.Struct.Biol., 9, 2002
|
|
1KHZ
 
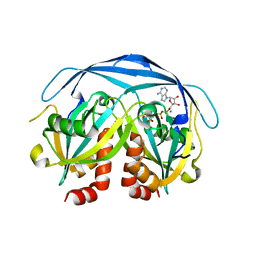 | | Structure of the ADPR-ase in complex with AMPCPR and Mg | | Descriptor: | ADP-ribose pyrophosphatase, ALPHA-BETA METHYLENE ADP-RIBOSE, CHLORIDE ION, ... | | Authors: | Gabelli, S.B, Bianchet, M.A, Bessman, M.J, Amzel, L.M. | | Deposit date: | 2001-12-01 | | Release date: | 2002-10-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Mechanism of the Escherichia coli ADP-ribose pyrophosphatase, a Nudix hydrolase.
Biochemistry, 41, 2002
|
|
1MQE
 
 | | Structure of the MT-ADPRase in complex with gadolidium and ADP-ribose, a Nudix enzyme | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, ADPR pyrophosphatase, GADOLINIUM ION | | Authors: | Kang, L.-W, Gabelli, S.B, Bianchet, M.A, Cunningham, J.E, O'Handley, S.F, Amzel, L.M. | | Deposit date: | 2002-09-16 | | Release date: | 2003-08-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and mechanism of MT-ADPRase, a Nudix hydrolase from Mycobacterium tuberculosis
Structure, 11, 2003
|
|
