1L1Y
 
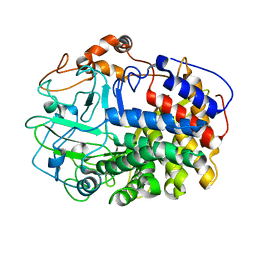 | | The Crystal Structure and Catalytic Mechanism of Cellobiohydrolase CelS, the Major Enzymatic Component of the Clostridium thermocellum cellulosome | | Descriptor: | beta-D-glucopyranose-(1-4)-beta-D-glucopyranose, cellobiohydrolase | | Authors: | Guimaraes, B.G, Souchon, H, Lytle, B.L, Wu, J.H.D, Alzari, P.M. | | Deposit date: | 2002-02-20 | | Release date: | 2002-07-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure and catalytic mechanism of cellobiohydrolase CelS, the major enzymatic component of the Clostridium thermocellum Cellulosome.
J.Mol.Biol., 320, 2002
|
|
1L2A
 
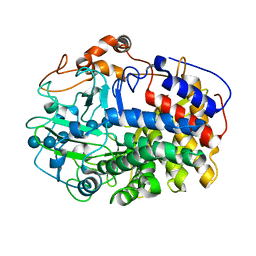 | | The Crystal Structure and Catalytic Mechanism of Cellobiohydrolase CelS, the Major Enzymatic Component of the Clostridium thermocellum cellulosome | | Descriptor: | beta-D-glucopyranose-(1-4)-beta-D-glucopyranose, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose, cellobiohydrolase | | Authors: | Guimaraes, B.G, Souchon, H, Lytle, B.L, Wu, J.H.D, Alzari, P.M. | | Deposit date: | 2002-02-20 | | Release date: | 2002-07-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure and catalytic mechanism of cellobiohydrolase CelS, the major enzymatic component of the Clostridium thermocellum Cellulosome.
J.Mol.Biol., 320, 2002
|
|
1IS9
 
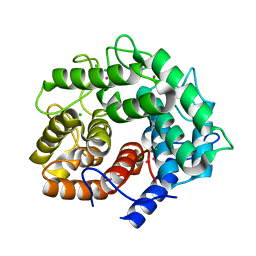 | | Endoglucanase A from Clostridium thermocellum at atomic resolution | | Descriptor: | CHLORIDE ION, MERCURY (II) ION, endoglucanase A | | Authors: | Schmidt, A, Gonzalez, A, Morris, R.J, Costabel, M, Alzari, P.M, Lamzin, V.S. | | Deposit date: | 2001-11-26 | | Release date: | 2002-09-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Advantages of high-resolution phasing: MAD to atomic resolution.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1JTG
 
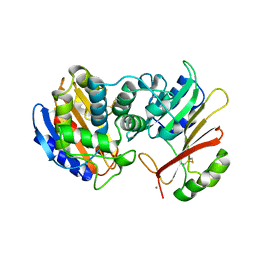 | | CRYSTAL STRUCTURE OF TEM-1 BETA-LACTAMASE / BETA-LACTAMASE INHIBITOR PROTEIN COMPLEX | | Descriptor: | BETA-LACTAMASE INHIBITORY PROTEIN, BETA-LACTAMASE TEM, CALCIUM ION | | Authors: | Strynadka, N.C.J, Jensen, S.E, Alzari, P.M, James, M.N. | | Deposit date: | 2001-08-20 | | Release date: | 2001-10-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal structure and kinetic analysis of beta-lactamase inhibitor protein-II in complex with TEM-1 beta-lactamase.
Nat.Struct.Biol., 8, 2001
|
|
2UYL
 
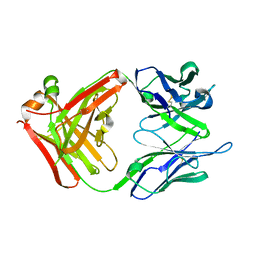 | | Crystal structure of a monoclonal antibody directed against an antigenic determinant common to Ogawa and Inaba serotypes of Vibrio cholerae O1 | | Descriptor: | MONOCLONAL ANTIBODY F-22-30 | | Authors: | Ahmed, F, Haouz, A, Nato, F, Fournier, J.M, Alzari, P.M. | | Deposit date: | 2007-04-10 | | Release date: | 2008-05-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of a Monoclonal Antibody Directed Against an Antigenic Determinant Common to Ogawa and Inaba Serotypes of Vibrio Cholerae O1.
Proteins, 70, 2008
|
|
2X2B
 
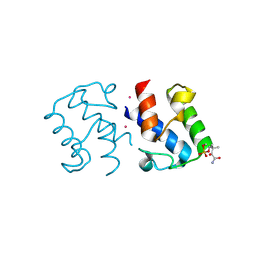 | | Crystal structure of malonyl-ACP (acyl carrier protein) from Bacillus subtilis | | Descriptor: | 3-{[2-({N-[(2S)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-beta-alanyl}amino)ethyl]sulfanyl}-3-oxopropanoic acid, ACYL CARRIER PROTEIN, PLATINUM (II) ION | | Authors: | Bellinzoni, M, Martinez, M.A, DeMendoza, D, Alzari, P.M. | | Deposit date: | 2010-01-12 | | Release date: | 2010-03-16 | | Last modified: | 2011-10-12 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | A Novel Role of Malonyl-Acp in Lipid Homeostasis.
Biochemistry, 49, 2010
|
|
2V06
 
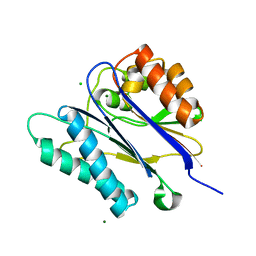 | | Crystal structure of the PPM Ser-Thr phosphatase MsPP from Mycobacterium smegmatis at pH 5.5 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, SER-THR PHOSPHATASE MSPP | | Authors: | Wehenkel, A, Bellinzoni, M, Schaeffer, F, Villarino, A, Alzari, P.M. | | Deposit date: | 2007-05-08 | | Release date: | 2007-10-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Structural and Binding Studies of the Three-Metal Center in Two Mycobacterial Ppm Ser/Thr Protein Phosphatases.
J.Mol.Biol., 374, 2007
|
|
2WZV
 
 | | Crystal structure of the FMN-dependent nitroreductase NfnB from Mycobacterium smegmatis | | Descriptor: | FLAVIN MONONUCLEOTIDE, GLYCEROL, NFNB PROTEIN, ... | | Authors: | Bellinzoni, M, Manina, G, Riccardi, G, Alzari, P.M. | | Deposit date: | 2009-12-03 | | Release date: | 2010-07-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Biological and Structural Characterization of the Mycobacterium Smegmatis Nitroreductase Nfnb, and its Role in Benzothiazinone Resistance
Mol.Microbiol., 77, 2010
|
|
2XIW
 
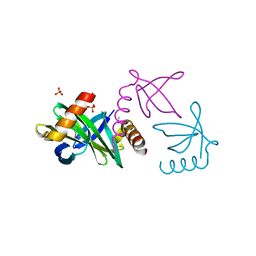 | | Crystal structure of the Sac7d-derived IgG1-binder C3-C24S | | Descriptor: | CHLORIDE ION, DNA-BINDING PROTEIN 7D, SULFATE ION | | Authors: | Bellinzoni, M, Colinet, S, Behar, G, Alzari, P.M, Pecorari, F. | | Deposit date: | 2010-07-01 | | Release date: | 2011-07-13 | | Last modified: | 2013-04-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Tolerance of the Archaeal Sac7D Scaffold Protein to Alternative Library Designs: Characterization of Anti-Immunoglobulin G Affitins.
Protein Eng.Des.Sel., 26, 2013
|
|
2V57
 
 | | Crystal structure of the TetR-like transcriptional regulator LfrR from Mycobacterium smegmatis in complex with proflavine | | Descriptor: | ISOPROPYL ALCOHOL, PROFLAVIN, SULFATE ION, ... | | Authors: | Bellinzoni, M, Buroni, S, Riccardi, G, De Rossi, E, Alzari, P.M. | | Deposit date: | 2008-10-02 | | Release date: | 2009-10-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Plasticity and Distinct Drug-Binding Modes of Lfrr, a Mycobacterial Efflux Pump Regulator.
J.Bacteriol., 191, 2009
|
|
2XT6
 
 | | Crystal structure of Mycobacterium smegmatis alpha-ketoglutarate decarboxylase homodimer (orthorhombic form) | | Descriptor: | 2-OXOGLUTARATE DECARBOXYLASE, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Wagner, T, Bellinzoni, M, Wehenkel, A.M, O'Hare, H.M, Alzari, P.M. | | Deposit date: | 2010-10-05 | | Release date: | 2011-06-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Functional Plasticity and Allosteric Regulation of Alpha-Ketoglutarate Decarboxylase in Central Mycobacterial Metabolism.
Chem.Biol., 18, 2011
|
|
2UYQ
 
 | | Crystal structure of ML2640c from Mycobacterium leprae in complex with S-adenosylmethionine | | Descriptor: | HYPOTHETICAL PROTEIN ML2640, S-ADENOSYLMETHIONINE | | Authors: | Grana, M, Buschiazzo, A, Wehenkel, A, Haouz, A, Miras, I, Shepard, W, Alzari, P.M. | | Deposit date: | 2007-04-11 | | Release date: | 2007-08-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Crystal Structure of M. Leprae Ml2640C Defines a Large Family of Putative S-Adenosylmethionine- Dependent Methyltransferases in Mycobacteria.
Protein Sci., 16, 2007
|
|
2UYO
 
 | | Crystal structure of ML2640c from Mycobacterium leprae in an hexagonal crystal form | | Descriptor: | HYPOTHETICAL PROTEIN ML2640 | | Authors: | Grana, M, Buschiazzo, A, Wehenkel, A, Haouz, A, Miras, I, Shepard, W, Alzari, P.M. | | Deposit date: | 2007-04-11 | | Release date: | 2007-08-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Crystal Structure of M. Leprae Ml2640C Defines a Large Family of Putative S-Adenosylmethionine- Dependent Methyltransferases in Mycobacteria.
Protein Sci., 16, 2007
|
|
2WAM
 
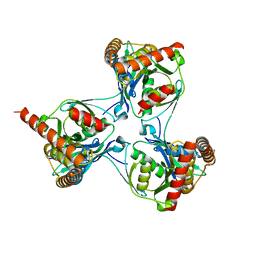 | | Crystal structure of Mycobacterium tuberculosis unknown function protein Rv2714 | | Descriptor: | CONSERVED HYPOTHETICAL ALANINE AND LEUCINE RICH PROTEIN, GLYCEROL | | Authors: | Bellinzoni, M, Grana, M, Buschiazzo, A, Miras, I, Haouz, A, Alzari, P.M. | | Deposit date: | 2009-02-09 | | Release date: | 2009-10-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of Mycobacterium Tuberculosis Rv2714, a Representative of a Duplicated Gene Family in Actinobacteria.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
2WGB
 
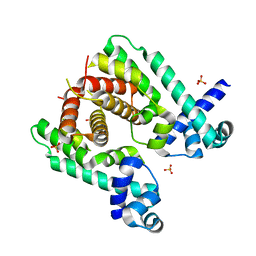 | | Crystal structure of the TetR-like transcriptional regulator LfrR from Mycobacterium smegmatis | | Descriptor: | SULFATE ION, TETR FAMILY TRANSCRIPTIONAL REPRESSOR LFRR | | Authors: | Bellinzoni, M, Buroni, S, Riccardi, G, De Rossi, E, Alzari, P.M. | | Deposit date: | 2009-04-16 | | Release date: | 2009-10-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Plasticity and Distinct Drug-Binding Modes of Lfrr, a Mycobacterial Efflux Pump Regulator.
J.Bacteriol., 191, 2009
|
|
2WZW
 
 | | Crystal structure of the FMN-dependent nitroreductase NfnB from Mycobacterium smegmatis in complex with NADPH | | Descriptor: | FLAVIN MONONUCLEOTIDE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NFNB PROTEIN, ... | | Authors: | Bellinzoni, M, Manina, G, Riccardi, G, Alzari, P.M. | | Deposit date: | 2009-12-03 | | Release date: | 2010-07-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Biological and Structural Characterization of the Mycobacterium Smegmatis Nitroreductase Nfnb, and its Role in Benzothiazinone Resistance
Mol.Microbiol., 77, 2010
|
|
2XTA
 
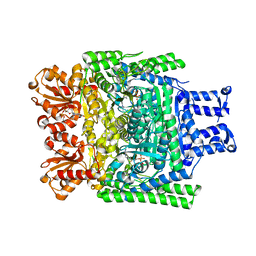 | | Crystal structure of the SucA domain of Mycobacterium smegmatis alpha- ketoglutarate decarboxylase in complex with acetyl-CoA (triclinic form) | | Descriptor: | 2-OXOGLUTARATE DECARBOXYLASE, ACETYL COENZYME *A, CALCIUM ION, ... | | Authors: | Wagner, T, Bellinzoni, M, Wehenkel, A.M, O'Hare, H.M, Alzari, P.M. | | Deposit date: | 2010-10-05 | | Release date: | 2011-06-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Functional Plasticity and Allosteric Regulation of Alpha-Ketoglutarate Decarboxylase in Central Mycobacterial Metabolism.
Chem.Biol., 18, 2011
|
|
2V7S
 
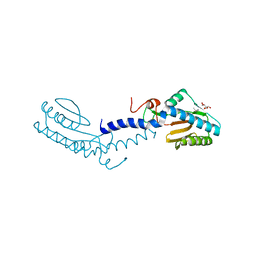 | | Crystal structure of the putative lipoprotein LppA from Mycobacterium tuberculosis | | Descriptor: | GLYCEROL, PROBABLE CONSERVED LIPOPROTEIN LPPA | | Authors: | Grana, M, Miras, I, Haouz, A, Winter, N, Buschiazzo, A, Bellinzoni, M, Alzari, P.M. | | Deposit date: | 2007-08-01 | | Release date: | 2008-08-26 | | Last modified: | 2011-09-28 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal Structure of Mycobacterium Tuberculosis Lppa, a Lipoprotein Confined to Pathogenic Mycobacteria.
Proteins, 78, 2010
|
|
2Y0P
 
 | | Crystal structure of the SucA domain of Mycobacterium smegmatis alpha- ketoglutarate decarboxylase in complex with the enamine-ThDP intermediate and acetyl-CoA | | Descriptor: | (4E)-4-{3-[(4-amino-2-methylpyrimidin-5-yl)methyl]-5-(2-{[(S)-hydroxy(phosphonooxy)phosphoryl]oxy}ethyl)-4-methyl-1,3-thiazol-2(3H)-ylidene}-4-hydroxybutanoic acid, 2-OXOGLUTARATE DECARBOXYLASE, ACETYL COENZYME *A, ... | | Authors: | Wagner, T, Bellinzoni, M, Wehenkel, A.M, O'Hare, H.M, Alzari, P.M. | | Deposit date: | 2010-12-07 | | Release date: | 2011-06-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Functional Plasticity and Allosteric Regulation of Alpha-Ketoglutarate Decarboxylase in Central Mycobacterial Metabolism.
Chem.Biol., 18, 2011
|
|
2Y7F
 
 | |
3ZHR
 
 | | Crystal structure of the H747A mutant of the SucA domain of Mycobacterium smegmatis KGD showing the active site lid closed | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Wagner, T, Barilone, N, Bellinzoni, M, Alzari, P.M. | | Deposit date: | 2012-12-24 | | Release date: | 2013-11-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Dual Conformation of the Post-Decarboxylation Intermediate is Associated with Distinct Enzyme States in Mycobacterial Alpha-Ketoglutarate Decarboxylase (Kgd).
Biochem.J., 457, 2014
|
|
4ADL
 
 | | Crystal structures of Rv1098c in complex with malate | | Descriptor: | (2S)-2-hydroxybutanedioic acid, FUMARATE HYDRATASE CLASS II | | Authors: | Mechaly, A.E, Haouz, A, Miras, I, Weber, P, Shepard, W, Cole, S, Alzari, P.M, Bellinzoni, M. | | Deposit date: | 2011-12-26 | | Release date: | 2012-04-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Conformational Changes Upon Ligand Binding in the Essential Class II Fumarase Rv1098C from Mycobacterium Tuberculosis.
FEBS Lett., 586, 2012
|
|
4A0Z
 
 | | Structure of the global transcription regulator FapR from Staphylococcus aureus in complex with malonyl-CoA | | Descriptor: | MALONYL-COENZYME A, TRANSCRIPTION FACTOR FAPR | | Authors: | Albanesi, D, Guerin, M.E, Buschiazzo, A, de Mendoza, D, Alzari, P.M. | | Deposit date: | 2011-09-13 | | Release date: | 2012-09-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for Feed-Forward Transcriptional Regulation of Membrane Lipid Homeostasis in Staphylococcus Aureus.
Plos Pathog., 9, 2013
|
|
4APA
 
 | |
4A0Y
 
 | | Structure of the global transcription regulator FapR from Staphylococcus aureus | | Descriptor: | CHLORIDE ION, TRANSCRIPTION FACTOR FAPR | | Authors: | Albanesi, D, Guerin, M.E, Buschiazzo, A, de Mendoza, D, Alzari, P.M. | | Deposit date: | 2011-09-13 | | Release date: | 2012-09-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis for Feed-Forward Transcriptional Regulation of Membrane Lipid Homeostasis in Staphylococcus Aureus.
Plos Pathog., 9, 2013
|
|
