4APB
 
 | | Crystal structure of Mycobacterium tuberculosis fumarase (Rv1098c) S318C in complex with fumarate | | Descriptor: | CALCIUM ION, FUMARATE HYDRATASE CLASS II, FUMARIC ACID | | Authors: | Bellinzoni, M, Haouz, A, Mechaly, A.E, Alzari, P.M. | | Deposit date: | 2012-03-31 | | Release date: | 2012-04-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Conformational Changes Upon Ligand Binding in the Essential Class II Fumarase Rv1098C from Mycobacterium Tuberculosis.
FEBS Lett., 586, 2012
|
|
4A12
 
 | | Structure of the global transcription regulator FapR from Staphylococcus aureus in complex with DNA operator | | Descriptor: | FAPR PROMOTER, TRANSCRIPTION FACTOR FAPR | | Authors: | Albanesi, D, Guerin, M.E, Buschiazzo, A, de Mendoza, D, Alzari, P.M. | | Deposit date: | 2011-09-13 | | Release date: | 2012-09-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural Basis for Feed-Forward Transcriptional Regulation of Membrane Lipid Homeostasis in Staphylococcus Aureus.
Plos Pathog., 9, 2013
|
|
4BIZ
 
 | |
4APA
 
 | |
3ZHT
 
 | | Crystal structure of the SucA domain of Mycobacterium smegmatis KGD, first post-decarboxylation intermediate from 2-oxoadipate | | Descriptor: | (5S)-5-{3-[(4-amino-2-methylpyrimidin-5-yl)methyl]-4-methyl-5-(2-{[(phosphonatooxy)phosphinato]oxy}ethyl)-1,3-thiazol-3-ium-2-yl}-5-hydroxypentanoate, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Wagner, T, Barilone, N, Bellinzoni, M, Alzari, P.M. | | Deposit date: | 2012-12-24 | | Release date: | 2013-11-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A Dual Conformation of the Post-Decarboxylation Intermediate is Associated with Distinct Enzyme States in Mycobacterial Alpha-Ketoglutarate Decarboxylase (Kgd).
Biochem.J., 457, 2014
|
|
3ZHU
 
 | | Crystal structure of the SucA domain of Mycobacterium smegmatis KGD, second post-decarboxylation intermediate from 2-oxoadipate | | Descriptor: | (5R)-5-{3-[(4-amino-2-methylpyrimidin-5-yl)methyl]-4-methyl-5-(2-{[(phosphonatooxy)phosphinato]oxy}ethyl)-1,3-thiazol-3-ium-2-yl}-5-hydroxypentanoate, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Wagner, T, Barilone, N, Bellinzoni, M, Alzari, P.M. | | Deposit date: | 2012-12-24 | | Release date: | 2013-11-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A Dual Conformation of the Post-Decarboxylation Intermediate is Associated with Distinct Enzyme States in Mycobacterial Alpha-Ketoglutarate Decarboxylase (Kgd).
Biochem.J., 457, 2014
|
|
3ZHS
 
 | | Crystal structure of the SucA domain of Mycobacterium smegmatis KGD, first post-decarboxylation intermediate from alpha-ketoglutarate | | Descriptor: | (4S)-4-{3-[(4-amino-2-methylpyrimidin-5-yl)methyl]-5-(2-{[(S)-hydroxy(phosphonooxy)phosphoryl]oxy}ethyl)-4-methyl-1,3lambda~5~-thiazol-2-yl}-4-hydroxybutanoic acid, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Wagner, T, Barilone, N, Bellinzoni, M, Alzari, P.M. | | Deposit date: | 2012-12-24 | | Release date: | 2013-11-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Dual Conformation of the Post-Decarboxylation Intermediate is Associated with Distinct Enzyme States in Mycobacterial Alpha-Ketoglutarate Decarboxylase (Kgd).
Biochem.J., 457, 2014
|
|
3ZHQ
 
 | | Crystal structure of the H747A mutant of the SucA domain of Mycobacterium smegmatis KGD | | Descriptor: | CALCIUM ION, MAGNESIUM ION, MULTIFUNCTIONAL 2-OXOGLUTARATE METABOLISM ENZYME, ... | | Authors: | Wagner, T, Barilone, N, Bellinzoni, M, Alzari, P.M. | | Deposit date: | 2012-12-24 | | Release date: | 2013-11-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Dual Conformation of the Post-Decarboxylation Intermediate is Associated with Distinct Enzyme States in Mycobacterial Alpha-Ketoglutarate Decarboxylase (Kgd).
Biochem.J., 457, 2014
|
|
3ZHV
 
 | | Crystal structure of the SucA domain of Mycobacterium smegmatis KGD, post-decarboxylation intermediate from pyruvate (2-hydroxyethyl-ThDP) | | Descriptor: | 2-[3-[(4-azanyl-2-methyl-pyrimidin-5-yl)methyl]-4-methyl-2-[(1S)-1-oxidanylethyl]-1,3-thiazol-3-ium-5-yl]ethyl phosphono hydrogen phosphate, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Wagner, T, Barilone, N, Bellinzoni, M, Alzari, P.M. | | Deposit date: | 2012-12-24 | | Release date: | 2013-11-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A Dual Conformation of the Post-Decarboxylation Intermediate is Associated with Distinct Enzyme States in Mycobacterial Alpha-Ketoglutarate Decarboxylase (Kgd).
Biochem.J., 457, 2014
|
|
1JTG
 
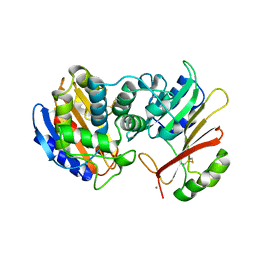 | | CRYSTAL STRUCTURE OF TEM-1 BETA-LACTAMASE / BETA-LACTAMASE INHIBITOR PROTEIN COMPLEX | | Descriptor: | BETA-LACTAMASE INHIBITORY PROTEIN, BETA-LACTAMASE TEM, CALCIUM ION | | Authors: | Strynadka, N.C.J, Jensen, S.E, Alzari, P.M, James, M.N. | | Deposit date: | 2001-08-20 | | Release date: | 2001-10-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal structure and kinetic analysis of beta-lactamase inhibitor protein-II in complex with TEM-1 beta-lactamase.
Nat.Struct.Biol., 8, 2001
|
|
4A0Y
 
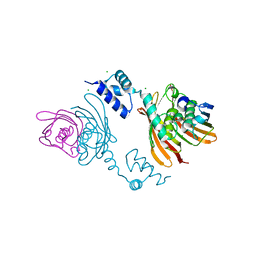 | | Structure of the global transcription regulator FapR from Staphylococcus aureus | | Descriptor: | CHLORIDE ION, TRANSCRIPTION FACTOR FAPR | | Authors: | Albanesi, D, Guerin, M.E, Buschiazzo, A, de Mendoza, D, Alzari, P.M. | | Deposit date: | 2011-09-13 | | Release date: | 2012-09-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis for Feed-Forward Transcriptional Regulation of Membrane Lipid Homeostasis in Staphylococcus Aureus.
Plos Pathog., 9, 2013
|
|
4AW1
 
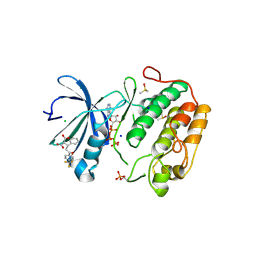 | | Human PDK1 Kinase Domain in Complex with Allosteric Compound PS210 Bound to the PIF-Pocket | | Descriptor: | 3-PHOSPHOINOSITIDE-DEPENDENT PROTEIN KINASE 1, ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Schulze, J.O, Busschots, K, Lopez-Garcia, L.A, Lammi, C, Stroba, A, Zeuzem, S, Piiper, A, Alzari, P.M, Neimanis, S, Arencibia, J.M, Engel, M, Biondi, R.M. | | Deposit date: | 2012-05-30 | | Release date: | 2012-10-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Substrate-Selective Inhibition of Protein Kinase Pdk1 by Small Compounds that Bind to the Pif-Pocket Allosteric Docking Site.
Chem.Biol., 19, 2012
|
|
4AW0
 
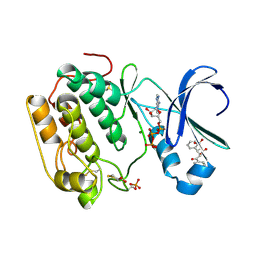 | | Human PDK1 Kinase Domain in Complex with Allosteric Compound PS182 Bound to the PIF-Pocket | | Descriptor: | 3-PHOSPHOINOSITIDE-DEPENDENT PROTEIN KINASE 1, ADENOSINE-5'-TRIPHOSPHATE, DIMETHYL SULFOXIDE, ... | | Authors: | Schulze, J.O, Busschots, K, Lopez-Garcia, L.A, Lammi, C, Stroba, A, Zeuzem, S, Piiper, A, Alzari, P.M, Neimanis, S, Arencibia, J.M, Engel, M, Biondi, R.M. | | Deposit date: | 2012-05-30 | | Release date: | 2012-10-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Substrate-Selective Inhibition of Protein Kinase Pdk1 by Small Compounds that Bind to the Pif-Pocket Allosteric Docking Site.
Chem.Biol., 19, 2012
|
|
4A0Z
 
 | | Structure of the global transcription regulator FapR from Staphylococcus aureus in complex with malonyl-CoA | | Descriptor: | MALONYL-COENZYME A, TRANSCRIPTION FACTOR FAPR | | Authors: | Albanesi, D, Guerin, M.E, Buschiazzo, A, de Mendoza, D, Alzari, P.M. | | Deposit date: | 2011-09-13 | | Release date: | 2012-09-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for Feed-Forward Transcriptional Regulation of Membrane Lipid Homeostasis in Staphylococcus Aureus.
Plos Pathog., 9, 2013
|
|
4A07
 
 | | Human PDK1 Kinase Domain in Complex with Allosteric Activator PS171 Bound to the PIF-Pocket | | Descriptor: | (3S)-3-(4-CHLOROPHENYL)-4-(5,7-DICHLORO-1H-BENZIMIDAZOL-2-YL)BUTANOIC ACID, 3-PHOSPHOINOSITIDE-DEPENDENT PROTEIN KINASE 1, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Schulze, J.O, Lopez-Garcia, L.A, Froehner, W, Zhang, H, Navratil, J, Hindie, V, Zeuzem, S, Alzari, P.M, Neimanis, S, Engel, M, Biondi, R.M. | | Deposit date: | 2011-09-08 | | Release date: | 2011-12-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Allosteric Regulation of Protein Kinase Pkczeta by the N-Terminal C1 Domain and Small Compounds to the Pif-Pocket.
Chem.Biol., 18, 2011
|
|
4A06
 
 | | Human PDK1 Kinase Domain in Complex with Allosteric Activator PS114 Bound to the PIF-Pocket | | Descriptor: | (3S)-4-(5-chloro-1H-benzimidazol-2-yl)-3-(4-chlorophenyl)butanoic acid, 3-PHOSPHOINOSITIDE-DEPENDENT PROTEIN KINASE 1, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Schulze, J.O, Lopez-Garcia, L.A, Froehner, W, Zhang, H, Navratil, J, Hindie, V, Zeuzem, S, Alzari, P.M, Neimanis, S, Engel, M, Biondi, R.M. | | Deposit date: | 2011-09-08 | | Release date: | 2011-12-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Allosteric Regulation of Protein Kinase Pkczeta by the N-Terminal C1 Domain and Small Compounds to the Pif-Pocket.
Chem.Biol., 18, 2011
|
|
4BIW
 
 | | Crystal structure of CpxAHDC (hexagonal form) | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, SENSOR PROTEIN CPXA, ... | | Authors: | Mechaly, A.E, Sassoon, N, Betton, J.M, Alzari, P.M. | | Deposit date: | 2013-04-13 | | Release date: | 2014-02-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Segmental Helical Motions and Dynamical Asymmetry Modulate Histidine Kinase Autophosphorylation.
Plos Biol., 12, 2014
|
|
4BIV
 
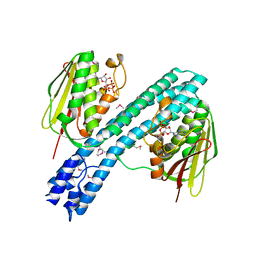 | | Crystal structure of CpxAHDC (trigonal form) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, SENSOR PROTEIN CPXA | | Authors: | Mechaly, A.E, Sassoon, N, Betton, J.M, Alzari, P.M. | | Deposit date: | 2013-04-13 | | Release date: | 2014-02-12 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Segmental Helical Motions and Dynamical Asymmetry Modulate Histidine Kinase Autophosphorylation.
Plos Biol., 12, 2014
|
|
4CB0
 
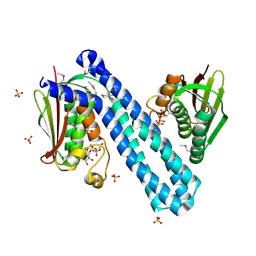 | | Crystal structure of CpxAHDC in complex with ATP (hexagonal form) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, SENSOR PROTEIN CPXA, SULFATE ION | | Authors: | Mechaly, A.E, Sassoon, N, Betton, J.M, Alzari, P.M. | | Deposit date: | 2013-10-09 | | Release date: | 2014-02-12 | | Last modified: | 2017-07-12 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Segmental Helical Motions and Dynamical Asymmetry Modulate Histidine Kinase Autophosphorylation.
Plos Biol., 12, 2014
|
|
4BIU
 
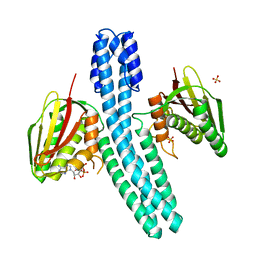 | | Crystal structure of CpxAHDC (orthorhombic form 1) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, SENSOR PROTEIN CPXA, SULFATE ION | | Authors: | Mechaly, A.E, Sassoon, N, Betton, J.M, Alzari, P.M. | | Deposit date: | 2013-04-13 | | Release date: | 2014-02-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | Segmental Helical Motions and Dynamical Asymmetry Modulate Histidine Kinase Autophosphorylation.
Plos Biol., 12, 2014
|
|
4BIX
 
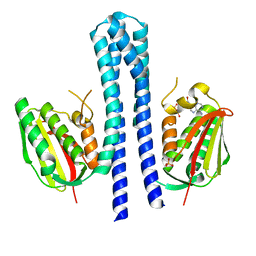 | | Crystal structure of CpxAHDC (monoclinic form 1) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, SENSOR PROTEIN CPXA, ... | | Authors: | Mechaly, A.E, Sassoon, N, Betton, J.M, Alzari, P.M. | | Deposit date: | 2013-04-13 | | Release date: | 2014-02-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Segmental Helical Motions and Dynamical Asymmetry Modulate Histidine Kinase Autophosphorylation.
Plos Biol., 12, 2014
|
|
4BIY
 
 | | Crystal structure of CpxAHDC (monoclinic form 2) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, SENSOR PROTEIN CPXA, SULFATE ION | | Authors: | Mechaly, A.E, Sassoon, N, Betton, J.M, Alzari, P.M. | | Deposit date: | 2013-04-13 | | Release date: | 2014-02-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Segmental Helical Motions and Dynamical Asymmetry Modulate Histidine Kinase Autophosphorylation.
Plos Biol., 12, 2014
|
|
4A0X
 
 | | Structure of the global transcription regulator FapR from Staphylococcus aureus | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ACETATE ION, TRANSCRIPTION FACTOR FAPR, ... | | Authors: | Albanesi, D, Guerin, M.E, Buschiazzo, A, de Mendoza, D, Alzari, P.M. | | Deposit date: | 2011-09-13 | | Release date: | 2012-09-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis for Feed-Forward Transcriptional Regulation of Membrane Lipid Homeostasis in Staphylococcus Aureus.
Plos Pathog., 9, 2013
|
|
