4QS5
 
 | | CRYSTAL STRUCTURE of 5-CARBOXYVANILLATE DECARBOXYLASE LIGW2 FROM NOVOSPHINGOBIUM AROMATICIVORANS DSM 12444 (TARGET EFI-505250) WITH BOUND MANGANESE AND 3-methoxy-4-hydroxy-5-nitrobenzoic acid, THE D314N MUTANT | | Descriptor: | 4-hydroxy-3-methoxy-5-nitrobenzoic acid, ACETATE ION, CHLORIDE ION, ... | | Authors: | Patskovsky, Y, Vladimirova, A, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Raushel, M, Almo, S.C. | | Deposit date: | 2014-07-02 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of 5-CARBOXYVANILLATE Decarboxylase from Novosphingobium Aromaticivorans
To be Published
|
|
4QRO
 
 | | CRYSTAL STRUCTURE of DIHYDROXYBENZOIC ACID DECARBBOXYLASE BPRO_2061 (TARGET EFI-500288) FROM POLAROMONAS SP. JS666 WITH BOUND MANGANESE AND AN INHIBITOR, 2-NITRORESORCINOL | | Descriptor: | 2-nitrobenzene-1,3-diol, ACETATE ION, BICARBONATE ION, ... | | Authors: | Patskovsky, Y, Vladimirova, A, Toro, R, Bhosle, R, Gerlt, J.A, Raushel, M, Almo, S.C. | | Deposit date: | 2014-07-01 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of Dihydroxybenzoate Decarboxylase from Frompolaromonas Sp WITH BOUND MANGANESE AND 2-NITRORESORCINOL
To be Published
|
|
4R2F
 
 | | Crystal structure of sugar transporter ACHL_0255 from Arthrobacter chlorophenolicus A6, target EFI-510633, with bound laminaribiose | | Descriptor: | Extracellular solute-binding protein family 1, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Al Obaidi, N, Chamala, S, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Lafleur, J, Siedel, R.D, Hillerich, B, Love, J, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-08-11 | | Release date: | 2014-08-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of sugar transporter ACHL_0255 from Arthrobacter chlorophenolicus, target EFI-510633
To be Published
|
|
4R6Y
 
 | | Crystal structure of solute-binding protein stm0429 from salmonella enterica subsp. enterica serovar typhimurium str. lt2, target efi-510776, a closed conformation, in complex with glycerol and acetate | | Descriptor: | ACETATE ION, GLYCEROL, Putative 2-aminoethylphosphonate-binding periplasmic protein | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Al obaidi, N, Chamala, S, Attonito, J.D, Scott glenn, A, Chowdhury, S, Lafleur, J, Siedel, R.D, Hillerich, B, Love, J, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-08-26 | | Release date: | 2014-09-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Crystal Structure of Transporter STM0429 from Salmonella Enterica
To be Published
|
|
4QSE
 
 | | Crystal structure of ATU4361 sugar transporter from Agrobacterium Fabrum c58, target efi-510558, with bound glycerol | | Descriptor: | ABC-TYPE SUGAR TRANSPORTER, GLYCEROL | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Al Obaidi, N, Chamala, S, Attonito, K.L, Scott Glenn, A, Chowdhury, S, Lafleur, J, Siedel, R.D, Hillerich, B, Love, J, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-07-03 | | Release date: | 2014-08-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Crystal structure of maltoside transporter ATU4361 from
Agrobacterium Fabrum, target EFI-510558
To be Published
|
|
4R88
 
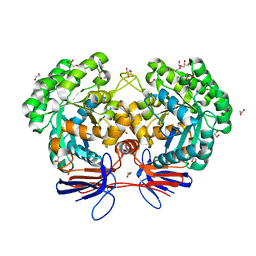 | | Crystal structure of 5-methylcytosine deaminase from Klebsiella pneumoniae liganded with 5-fluorocytosine | | Descriptor: | 1,2-ETHANEDIOL, 5-fluorocytosine, ACETIC ACID, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Hitchcock, D.S, Raushel, F.M, Almo, S.C. | | Deposit date: | 2014-08-29 | | Release date: | 2014-09-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.952 Å) | | Cite: | Crystal structure of 5-methylcytosine deaminase from Klebsiella pneumoniae liganded with 5-fluorocytosine
To be Published
|
|
4QUP
 
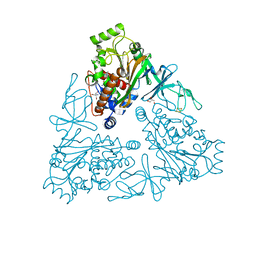 | | Crystal structure of stachydrine demethylase with N-methyl proline from low X-ray dose composite datasets | | Descriptor: | 1-methyl-L-proline, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, COBALT HEXAMMINE(III), ... | | Authors: | Agarwal, R, Andi, B, Gizzi, A, Bonanno, J.B, Almo, S.C, Orville, A.M. | | Deposit date: | 2014-07-11 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Tracking photoelectron induced in-crystallo enzyme catalysis
To be Published
|
|
4R31
 
 | |
4QSC
 
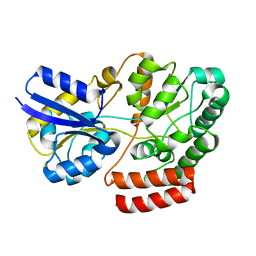 | | Crystal structure of ATU4361 sugar transporter from Agrobacterium Fabrum C58, target efi-510558, with bound maltose | | Descriptor: | ABC-TYPE SUGAR TRANSPORTER, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Al Obaidi, N, Chamala, S, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Lafleur, J, Siedel, R.D, Hillerich, B, Love, J, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-07-03 | | Release date: | 2014-07-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal Structure of Maltoside Transporter Atu4361 from Agrobacterium Fabrum, Target Efi-510558
To be Published
|
|
4RDV
 
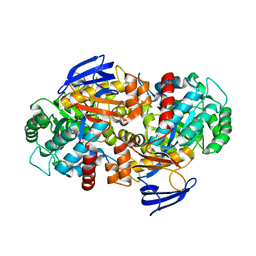 | | The structure of N-formimino-L-Glutamate Iminohydrolase from Pseudomonas aeruginosa complexed with N-formimino-L-Aspartate | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, N-FORMIMINO-L-GLUTAMATE IMINOHYDROLASE, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Marti-Arbona, R, Raushel, F.M, Almo, S.C. | | Deposit date: | 2014-09-19 | | Release date: | 2014-10-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | The structure of N-formimino-L-Glutamate Iminohydrolase from Pseudomonas aeruginosa complexed with N-formimino-L-Aspartate
To be Published
|
|
4QRZ
 
 | | Crystal structure of sugar transporter atu4361 from agrobacterium fabrum c58, target efi-510558, with bound maltotriose | | Descriptor: | ABC-TYPE SUGAR TRANSPORTER, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Al Obaidi, N, Stead, M, Washington, E, Glenn, A.S, Chowdhury, S, Evans, B, Hammonds, J, Love, J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-07-02 | | Release date: | 2014-07-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Crystal Structure of Maltoside Transporter from Agrobacterium Radiobacter, Target Efi-510558
To be Published
|
|
4QSD
 
 | | Crystal structure of atu4361 sugar transporter from Agrobacterium Fabrum C58, target efi-510558, with bound sucrose | | Descriptor: | ABC-TYPE SUGAR TRANSPORTER, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Al Obaidi, N, Chamala, S, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Lafleur, J, Siedel, R.D, Hillerich, B, Love, J, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-07-03 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Crystal Structure of Maltoside Transporter Atu4361 from Agrobacterium Fabrum, Target Efi-510558
To be Published
|
|
4QTG
 
 | | CRYSTAL STRUCTURE of 5-CARBOXYVANILLATE DECARBOXYLASE LIGW2 (TARGET EFI-505250) FROM NOVOSPHINGOBIUM AROMATICIVORANS DSM 12444 COMPLEXED WITH MANGANESE | | Descriptor: | 5-CARBOXYVANILLATE DECARBOXYLASE, GLYCEROL, MANGANESE (II) ION | | Authors: | Patskovsky, Y, Vladimirova, A, Toro, R, Bhosle, R, Gerlt, J.A, Raushel, F.M, Almo, S.C. | | Deposit date: | 2014-07-07 | | Release date: | 2014-08-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Crystal Structure of 5-Carboxyvanillate Decarboxylase LIGW2 from Novosphingobium Aromaticivorans
To be Published
|
|
4R2B
 
 | | Crystal structure of sugar transporter Oant_3817 from Ochrobactrum anthropi, target EFI-510528, with bound glucose | | Descriptor: | Extracellular solute-binding protein family 1, alpha-D-glucopyranose | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Al Obaidi, N, Chamala, S, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Lafleur, J, Siedel, R.D, Hillerich, B, Love, J, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-08-11 | | Release date: | 2014-08-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystal Structure of Glucose Transporter Oant_3817 from Ochrobactrum Anthropi, Target EFI-510528
To be Published
|
|
4QSF
 
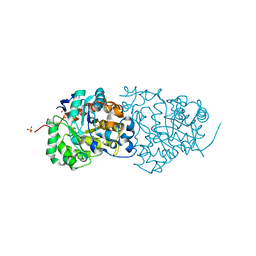 | | CRYSTAL STRUCTURE of AMIDOHYDROLASE PMI1525 (TARGET EFI-500319) FROM PROTEUS MIRABILIS HI4320, A COMPLEX WITH BUTYRIC ACID AND MANGANESE | | Descriptor: | Amidohydrolase Pmi1525, MANGANESE (II) ION, SULFATE ION, ... | | Authors: | Patskovsky, Y, Toro, R, Xiang, D.F, Raushel, F.M, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-07-03 | | Release date: | 2014-07-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of Amidohydrolase Pmi1525 from Proteus Mirabilis Hi4320
To be Published
|
|
4R85
 
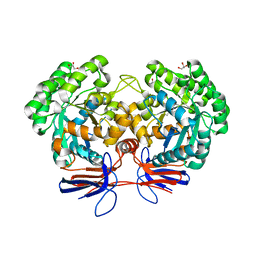 | | Crystal structure of 5-methylcytosine deaminase from Klebsiella pneumoniae liganded with 5-methylcytosine | | Descriptor: | 5-methylcytosine, Cytosine deaminase, FE (II) ION, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Hitchcock, D.S, Raushel, F.M, Almo, S.C. | | Deposit date: | 2014-08-29 | | Release date: | 2014-09-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Crystal structure of 5-methylcytosine deaminase from Klebsiella pneumoniae liganded with 5-methylcytosine
To be Published
|
|
4QUR
 
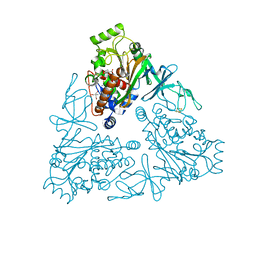 | | Crystal Structure of stachydrine demethylase in complex with cyanide, oxygen, and N-methyl proline in a new orientation | | Descriptor: | 1-methyl-L-proline, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, COBALT HEXAMMINE(III), ... | | Authors: | Agarwal, R, Andi, B, Gizzi, A, Bonanno, J.B, Almo, S.C, Orville, A.M. | | Deposit date: | 2014-07-11 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.759 Å) | | Cite: | Tracking photoelectron induced in-crystallo enzyme catalysis
To be Published
|
|
4R6H
 
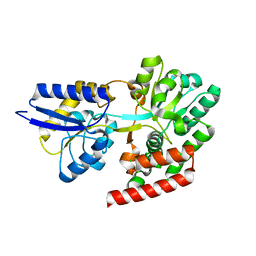 | | Crystal structure of putative binding protein msme from bacillus subtilis subsp. subtilis str. 168, target efi-510764, an open conformation | | Descriptor: | CHLORIDE ION, Solute binding protein MsmE | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Al obaidi, N, Chamala, S, Attonito, J.D, Scott glenn, A, Chowdhury, S, Lafleur, J, Siedel, R.D, Hillerich, B, Love, J, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-08-25 | | Release date: | 2014-09-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of Transporter Msme from Bacillus Subtilis, Target Efi-510764
To be Published
|
|
4R7W
 
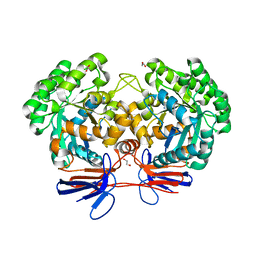 | | Crystal structure of 5-methylcytosine deaminase from Klebsiella pneumoniae liganded with phosphonocytosine | | Descriptor: | (2R)-2-amino-2,5-dihydro-1,5,2-diazaphosphinin-6(1H)-one 2-oxide, 1,2-ETHANEDIOL, Cytosine deaminase, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Hitchcock, D.S, Raushel, F.M, Almo, S.C. | | Deposit date: | 2014-08-28 | | Release date: | 2014-09-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Crystal structure of 5-methylcytosine deaminase from Klebsiella pneumoniae liganded with phosphonocytosine
To be Published
|
|
4RDW
 
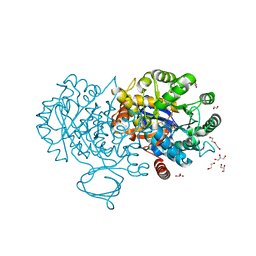 | | The structure of N-formimino-L-Glutamate Iminohydrolase from Pseudomonas aeruginosa complexed with N-Guanidino-L-Glutaric acid | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Marti-Arbona, R, Raushel, F.M, Almo, S.C. | | Deposit date: | 2014-09-19 | | Release date: | 2014-10-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.591 Å) | | Cite: | The structure of N-formimino-L-Glutamate Iminohydrolase from Pseudomonas aeruginosa complexed with N-Guanidino-L-Glutaric acid
To be Published
|
|
4R6K
 
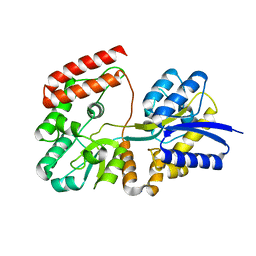 | | Crystal structure of ABC transporter substrate-binding protein YesO from Bacillus subtilis, TARGET EFI-510761, an open conformation | | Descriptor: | SODIUM ION, SOLUTE-BINDING PROTEIN | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Al Obaidi, N, Chamala, S, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Lafleur, J, Siedel, R.D, Hillerich, B, Love, J, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-08-25 | | Release date: | 2014-09-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal structure of transporter Yeso from Bacillus subtilis, Target Efi-510761
To be Published
|
|
4RXU
 
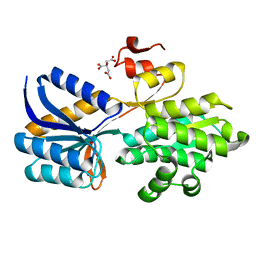 | | Crystal structure of carbohydrate transporter solute binding protein CAUR_1924 from Chloroflexus aurantiacus, Target EFI-511158, in complex with D-glucose | | Descriptor: | CITRIC ACID, Periplasmic sugar-binding protein, beta-D-glucopyranose | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Al Obaidi, N, Morisco, L.L, Wasserman, S.R, Chamala, S, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Lafleur, J, Hillerich, B, Siedel, R.D, Love, J, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-12-11 | | Release date: | 2014-12-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of sugar transporter CAUR_1924 from Chloroflexus aurantiacus, Target EFI-511158
To be Published
|
|
4RY1
 
 | | Crystal structure of periplasmic solute binding protein ECA2210 from Pectobacterium atrosepticum SCRI1043, Target EFI-510858 | | Descriptor: | ACETATE ION, GLYCEROL, Periplasmic solute binding protein | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Al Obaidi, N, Morisco, L.L, Wasserman, S.R, Chamala, S, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Lafleur, J, Hillerich, B, Siedel, R.D, Love, J, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-12-12 | | Release date: | 2014-12-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of periplasmic solute binding protein ECA2210 from Pectobacterium atrosepticum, Target EFI-510858
To be Published
|
|
4RWH
 
 | | Crystal structure of T cell costimulatory ligand B7-1 (CD80) | | Descriptor: | T-lymphocyte activation antigen CD80 | | Authors: | Fedorov, A.A, Fedorov, E.V, Samanta, D, Hillerich, B, Seidel, R.D, Almo, S.C. | | Deposit date: | 2014-12-04 | | Release date: | 2014-12-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Crystal structure of T cell costimulatory ligand B7-1 (CD80)
To be Published
|
|
4RK1
 
 | | Crystal structure of LacI family transcriptional regulator from Enterococcus faecium, Target EFI-512930, with bound ribose | | Descriptor: | CHLORIDE ION, Ribose transcriptional regulator, alpha-D-ribofuranose | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Al Obaidi, N, Chamala, S, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Lafleur, J, Siedel, R.D, Hillerich, B, Love, J, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-10-11 | | Release date: | 2014-10-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of LacI Transcriptional Regulator Rbsr from Enterococcus faecium, Target EFI-512930
To be Published
|
|
