1JKZ
 
 | | NMR Solution Structure of Pisum sativum defensin 1 (Psd1) | | Descriptor: | DEFENSE-RELATED PEPTIDE 1 | | Authors: | Almeida, M.S, Cabral, K.M.S, Kurtenbach, E, Almeida, F.C.L, Valente, A.P. | | Deposit date: | 2001-07-13 | | Release date: | 2002-02-06 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Pisum sativum defensin 1 by high resolution NMR: plant defensins, identical backbone with different mechanisms of action.
J.Mol.Biol., 315, 2002
|
|
1WCJ
 
 | | Conserved Hypothetical Protein TM0487 from Thermotoga maritima | | Descriptor: | HYPOTHETICAL PROTEIN TM0487 | | Authors: | Almeida, M.S, Peti, W, Herrmann, T, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2004-11-17 | | Release date: | 2004-12-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the Conserved Hypothetical Protein Tm0487 from Thermotoga Maritima: Implications for 216 Homologous Duf59 Proteins.
Protein Sci., 14, 2005
|
|
1UWD
 
 | | NMR STRUCTURE OF A PROTEIN WITH UNKNOWN FUNCTION FROM THERMOTOGA MARITIMA (TM0487), WHICH BELONGS TO THE DUF59 FAMILY. | | Descriptor: | HYPOTHETICAL PROTEIN TM0487 | | Authors: | Almeida, M.S, Peti, W, Herrmann, T, Wuthrich, K. | | Deposit date: | 2004-02-03 | | Release date: | 2004-12-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the Conserved Hypothetical Protein Tm0487 from Thermotoga Maritima: Implications for 216 Homologous Duf59 Proteins.
Protein Sci., 14, 2005
|
|
2HSX
 
 | | NMR Structure of the nonstructural protein 1 (nsp1) from the SARS coronavirus | | Descriptor: | Leader protein; p65 homolog; NSP1 (EC 3.4.22.-) | | Authors: | Almeida, M.S, Herrmann, T, Geralt, M, Johnson, M.A, Saikatendu, K, Joseph, J, Subramanian, R.C, Neuman, B.W, Buchmeier, M.J, Stevens, R.C, Kuhn, P, Wilson, I.A, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2006-07-24 | | Release date: | 2007-02-06 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Novel beta-barrel fold in the nuclear magnetic resonance structure of the replicase nonstructural protein 1 from the severe acute respiratory syndrome coronavirus.
J.Virol., 81, 2007
|
|
2GDT
 
 | | NMR Structure of the nonstructural protein 1 (nsp1) from the SARS coronavirus | | Descriptor: | Leader protein; p65 homolog; NSP1 (EC 3.4.22.-) | | Authors: | Almeida, M.S, Herrmann, T, Geralt, M, Johnson, M.A, Saikatendu, K, Joseph, J, Subramanian, R.C, Neuman, B.W, Buchmeier, M.J, Stevens, R.C, Kuhn, P, Wilson, I.A, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2006-03-17 | | Release date: | 2007-02-06 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Novel beta-barrel fold in the nuclear magnetic resonance structure of the replicase nonstructural protein 1 from the severe acute respiratory syndrome coronavirus.
J.Virol., 81, 2007
|
|
2LPN
 
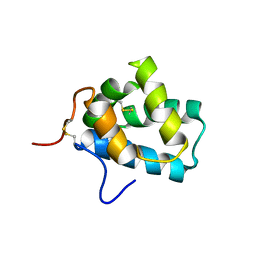 | | Solution Structure of N-Terminal domain of human Conserved Dopamine Neurotrophic Factor (CDNF) | | Descriptor: | Cerebral dopamine neurotrophic factor | | Authors: | Latge, C, Cabral, K.M.S, Foguel, D, Pires, J.R.M, Almeida, M.S. | | Deposit date: | 2012-02-15 | | Release date: | 2013-02-20 | | Last modified: | 2013-05-22 | | Method: | SOLUTION NMR | | Cite: | (1)H-, (13)C- and (15)N-NMR assignment of the N-terminal domain of human cerebral dopamine neurotrophic factor (CDNF).
Biomol.Nmr Assign., 7, 2013
|
|
2RLJ
 
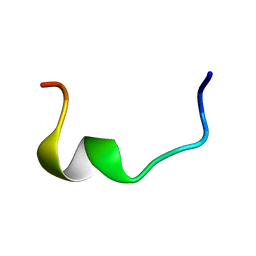 | | NMR Structure of Ebola fusion peptide in SDS micelles at pH 7 | | Descriptor: | Envelope glycoprotein | | Authors: | Freitas, M.S, Gaspar, L.P, Lorenzoni, M, Almeida, F.C, Tinoco, L.W, Almeida, M.S, Maia, L.F, Degreve, L, Valente, A.P, Silva, J.L. | | Deposit date: | 2007-07-05 | | Release date: | 2007-08-07 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of the Ebola fusion peptide in a membrane-mimetic environment and the interaction with lipid rafts.
J.Biol.Chem., 282, 2007
|
|
4BIT
 
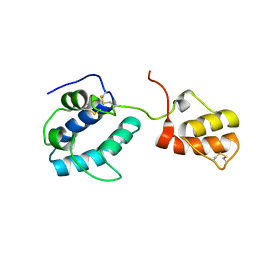 | | solution structure of cerebral dopamine neurotrophic factor (CDNF) | | Descriptor: | CEREBRAL DOPAMINE NEUROTROPHIC FACTOR | | Authors: | Latge, C, Cabral, K.M.S, Raymundo, D.P, Foguel, D, Herrmann, T, Almeida, M.S. | | Deposit date: | 2013-04-13 | | Release date: | 2014-04-23 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The Solution Structure and Dynamics of Full-Length Human Cerebral Dopamine Neurotrophic Factor and its Neuroprotective Role Against Alpha-Synuclein Oligomers.
J.Biol.Chem., 290, 2015
|
|
6UOG
 
 | | Asparaginase II from Escherichia coli | | Descriptor: | ASPARTIC ACID, L-asparaginase 2 | | Authors: | Araujo, T.S, Almeida, M.S, Lima, L.M.T.R. | | Deposit date: | 2019-10-14 | | Release date: | 2020-10-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Biophysical characterization of two commercially available preparations of the drug containing Escherichia coli L-Asparaginase 2.
Biophys.Chem., 271, 2021
|
|
6UOD
 
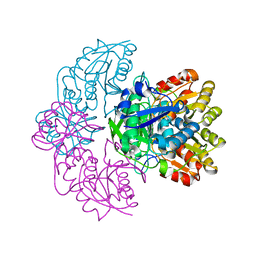 | | Asparaginase II from Escherichia coli | | Descriptor: | L-asparaginase 2 | | Authors: | Araujo, T.S, Almeida, M.S, Lima, L.M.T.R. | | Deposit date: | 2019-10-14 | | Release date: | 2020-10-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Biophysical characterization of two commercially available preparations of the drug containing Escherichia coli L-Asparaginase 2.
Biophys.Chem., 271, 2021
|
|
6UOH
 
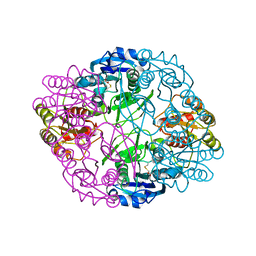 | | Asparaginase II from Escherichia coli | | Descriptor: | ASPARTIC ACID, L-asparaginase 2 | | Authors: | Araujo, T.S, Almeida, M.S, Lima, L.M.T.R. | | Deposit date: | 2019-10-14 | | Release date: | 2020-10-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Biophysical characterization of two commercially available preparations of the drug containing Escherichia coli L-Asparaginase 2.
Biophys.Chem., 271, 2021
|
|
2IDY
 
 | | NMR Structure of the SARS-CoV non-structural protein nsp3a | | Descriptor: | NSP3 | | Authors: | Serrano, P, Almeida, M.S, Johnson, M.A, Horst, R, Herrmann, T, Joseph, J, Saikatendu, K, Subramanian, V, Stevens, R.C, Kuhn, P, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2006-09-15 | | Release date: | 2006-12-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance structure of the N-terminal domain of nonstructural protein 3 from the severe acute respiratory syndrome coronavirus.
J.Virol., 81, 2007
|
|
2GRI
 
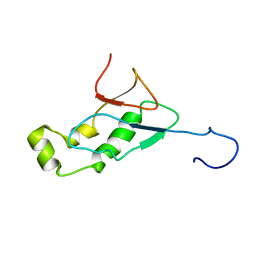 | | NMR Structure of the SARS-CoV non-structural protein nsp3a | | Descriptor: | NSP3 | | Authors: | Serrano, P, Almeida, M.S, Johnson, M.A, Herrmann, T, Saikatendu, K.S, Joseph, J, Subramanian, V, Neuman, B.W, Buchmeier, M.J, Stevens, R.C, Kuhn, P, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2006-04-24 | | Release date: | 2006-12-19 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance structure of the N-terminal domain of nonstructural protein 3 from the severe acute respiratory syndrome coronavirus.
J.Virol., 81, 2007
|
|
2I3B
 
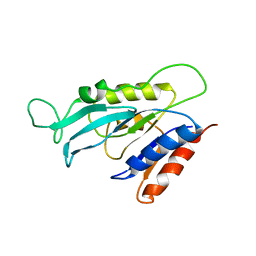 | |
