1FDM
 
 | |
1JKZ
 
 | | NMR Solution Structure of Pisum sativum defensin 1 (Psd1) | | Descriptor: | DEFENSE-RELATED PEPTIDE 1 | | Authors: | Almeida, M.S, Cabral, K.M.S, Kurtenbach, E, Almeida, F.C.L, Valente, A.P. | | Deposit date: | 2001-07-13 | | Release date: | 2002-02-06 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Pisum sativum defensin 1 by high resolution NMR: plant defensins, identical backbone with different mechanisms of action.
J.Mol.Biol., 315, 2002
|
|
9COJ
 
 | | SH3-like tandem domain of human KIN protein | | Descriptor: | DNA/RNA-binding protein KIN17 | | Authors: | de Lourenco, I.O, Fossey, M.A, Souza, F.P, Seixas, F.A.V, Fernandez, M.A, Almeida, F.C.L, Caruso, I.P. | | Deposit date: | 2024-07-16 | | Release date: | 2024-07-31 | | Method: | SOLUTION NMR | | Cite: | Solution structure of SH3-like tandem domain of human KIN protein and its interaction with RNA homo-oligonucleotides
To Be Published
|
|
6ALK
 
 | | NMR solution structure of the major beech pollen allergen Fag s 1 | | Descriptor: | Fag s 1 pollen allergen | | Authors: | Moraes, A.H, Asam, A, Almeida, F.C.L, Wallner, M, Ferreira, F, Valente, A.P. | | Deposit date: | 2017-08-08 | | Release date: | 2018-08-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for cross-reactivity and conformation fluctuation of the major beech pollen allergen Fag s 1.
Sci Rep, 8, 2018
|
|
8S9E
 
 | | Solution structure of jarastatin (rJast), a disintegrin from Bothrops jararaca | | Descriptor: | Disintegrin jarastatin | | Authors: | Vasconcelos, A.A, Estrada, J.E.C, Zingali, R.B, Almeida, F.C.L. | | Deposit date: | 2023-03-28 | | Release date: | 2023-11-29 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Toward the mechanism of jarastatin (rJast) inhibition of the integrin alpha V beta 3.
Int.J.Biol.Macromol., 255, 2024
|
|
5SZW
 
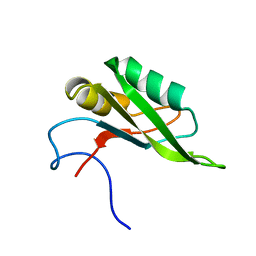 | | NMR solution structure of the RRM1 domain of the post-transcriptional regulator HuR | | Descriptor: | ELAV-like protein 1 | | Authors: | Lixa, C, Mujo, A, Jendiroba, K.A, Almeida, F.C.L, Lima, L.M.T.R, Pinheiro, A.S. | | Deposit date: | 2016-08-15 | | Release date: | 2017-09-06 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Oligomeric transition and dynamics of RNA binding by the HuR RRM1 domain in solution.
J. Biomol. NMR, 72, 2018
|
|
8EOD
 
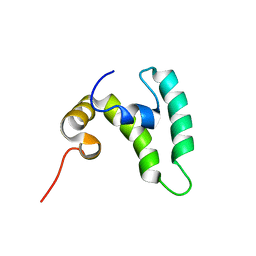 | |
7N45
 
 | |
8TG0
 
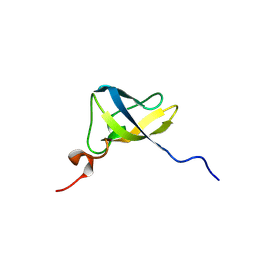 | |
6VK2
 
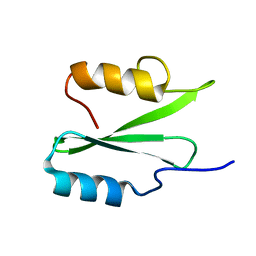 | |
1S4H
 
 | | NMR structure of cross-reactive peptides from L. braziliensis | | Descriptor: | 60S acidic ribosomal protein P2 | | Authors: | Soares, M.R, Bisch, P.M, Campos de Carvalho, A.C, Valente, A.P, Almeida, F.C.L. | | Deposit date: | 2004-01-16 | | Release date: | 2004-03-16 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Correlation between conformation and antibody binding: NMR structure of cross-reactive peptides from T. cruzi, human and L. braziliensis
Febs Lett., 560, 2004
|
|
1S4J
 
 | | NMR structure of cross-reactive peptides from Homo sapiens | | Descriptor: | 60S acidic ribosomal protein P2 | | Authors: | Soares, M.R, Bisch, P.M, Campos de Carvalho, A.C, Valente, A.P, Almeida, F.C.L. | | Deposit date: | 2004-01-16 | | Release date: | 2004-03-16 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Correlation between conformation and antibody binding: NMR structure of cross-reactive peptides from T. cruzi, human and L. braziliensis
Febs Lett., 560, 2004
|
|
6C44
 
 | | Zika virus capsid protein | | Descriptor: | Capsid protein | | Authors: | Morando, M.A, Barbosa, G.M, Cruz-Oliveira, C, Da Poian, A.T, Almeida, F.C.L. | | Deposit date: | 2018-01-11 | | Release date: | 2019-01-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Dynamics of Zika Virus Capsid Protein in Solution: The Properties and Exposure of the Hydrophobic Cleft Are Controlled by the alpha-Helix 1 Sequence.
Biochemistry, 58, 2019
|
|
6D6X
 
 | | HSP40 co-chaperone Sis1 J-domain | | Descriptor: | Type II HSP40 co-chaperone | | Authors: | Pinheiro, G.M.S, Amorim, G.C, Iqbal, A, Ramos, C.H.I, Almeida, F.C.L. | | Deposit date: | 2018-04-23 | | Release date: | 2019-05-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR investigation on the structure and function of the isolated J-domain from Sis1: Evidence of transient inter-domain interactions in the full-length protein.
Arch.Biochem.Biophys., 669, 2019
|
|
6NK9
 
 | |
6NOM
 
 | | NMR solution structure of Pisum sativum defensin 2 (Psd2) provides evidence for the presence of hydrophobic surface clusters | | Descriptor: | Defensin-2 | | Authors: | Pinheiro-Aguiar, R, Amaral, V.S.G, Bastos, I, Kurtenbach, E, Almeida, F.C.L. | | Deposit date: | 2019-01-16 | | Release date: | 2019-08-21 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance solution structure of Pisum sativum defensin 2 provides evidence for the presence of hydrophobic surface-clusters.
Proteins, 88, 2020
|
|
6NAN
 
 | | NMR structure determination of Ixolaris and Factor X interaction reveals a noncanonical mechanism of Kunitz inhibition | | Descriptor: | Ixolaris | | Authors: | De Paula, V.S, Sgourakis, N.G, Francischetti, I.M.B, Almeida, F.C.L, Monteiro, R.Q, Valente, A.P. | | Deposit date: | 2018-12-06 | | Release date: | 2019-06-12 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | NMR structure determination of Ixolaris and factor X(a) interaction reveals a noncanonical mechanism of Kunitz inhibition.
Blood, 134, 2019
|
|
6WBU
 
 | |
2HSY
 
 | |
2K9B
 
 | | Structure and membrane interactions of the antibiotic peptide dermadistinctin K by multidimensional solution and oriented 15N and 31P solid-state NMR spectroscopy | | Descriptor: | Dermadistinctin-K | | Authors: | Moraes, C.M, Verly, R.M, Resende, J.M, Aisenbrey, C, Bemquerer, M.P, Pilo-Veloso, D, Valente, A, Almeida, F.C.L, Bechinger, B. | | Deposit date: | 2008-10-07 | | Release date: | 2009-04-14 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Structure and membrane interactions of the antibiotic peptide dermadistinctin K by multidimensional solution and oriented 15N and 31P solid-state NMR spectroscopy.
Biophys.J., 96, 2009
|
|
2JX6
 
 | | Structure and membrane interactions of the antibiotic peptide dermadistinctin k by solution and oriented 15N and 31P solid-state NMR spectroscopy | | Descriptor: | Dermadistinctin-K | | Authors: | Mendonca Moraes, C, Verly, R.M, Resende, J.M, Bemquerer, M.P, Pilo-Veloso, D, Valente, A, Almeida, F.C.L, Bechinger, B. | | Deposit date: | 2007-11-08 | | Release date: | 2008-11-11 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Structure and membrane interactions of the antibiotic peptide dermadistinctin K by multidimensional solution and oriented 15N and 31P solid-state NMR spectroscopy
Biophys.J., 96, 2009
|
|
2I9H
 
 | |
2MBX
 
 | | Structure, dynamics and stability of allergen cod parvalbumin Gad m 1 by solution and high-pressure NMR. | | Descriptor: | CALCIUM ION, Parvalbumin beta | | Authors: | Moraes, A.H, Ackerbauer, D, Bublin, M, Ferreira, F, Almeida, F.C.L, Breiteneder, H, Valente, A. | | Deposit date: | 2013-08-07 | | Release date: | 2014-08-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution and high-pressure NMR studies of the structure, dynamics, and stability of the cross-reactive allergenic cod parvalbumin Gad m 1.
Proteins, 82, 2014
|
|
2LWL
 
 | | Structural Basis for the Interaction of Human β-Defensin 6 and Its Putative Chemokine Receptor CCR2 and Breast Cancer Microvesicles | | Descriptor: | Beta-defensin 106 | | Authors: | de Paula, V.S, Gomes, N.S.F, Lima, L.G, Miyamoto, C.A, Monteiro, R.Q, Almeida, F.C.L, Valente, A. | | Deposit date: | 2012-08-02 | | Release date: | 2013-08-21 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for the Interaction of Human beta-Defensin 6 and Its Putative Chemokine Receptor CCR2 and Breast Cancer Microvesicles.
J.Mol.Biol., 425, 2013
|
|
2M85
 
 | | PHD Domain from Human SHPRH | | Descriptor: | E3 ubiquitin-protein ligase SHPRH, ZINC ION | | Authors: | Machado, L.E.S.F, Pustovalova, Y, Pozhidaeva, A, Almeida, F.C.L, Bezsonova, I, Korzhnev, D.M. | | Deposit date: | 2013-05-07 | | Release date: | 2013-08-14 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | PHD domain from human SHPRH.
J.Biomol.Nmr, 56, 2013
|
|
