3L8G
 
 | | Crystal Structure of D,D-heptose 1.7-bisphosphate phosphatase from E. Coli complexed with D-glycero-D-manno-heptose 1 ,7-bisphosphate | | Descriptor: | 1,7-di-O-phosphono-L-glycero-beta-D-manno-heptopyranose, D,D-heptose 1,7-bisphosphate phosphatase, MAGNESIUM ION, ... | | Authors: | Nguyen, H, Peisach, E, Allen, K.N. | | Deposit date: | 2009-12-31 | | Release date: | 2010-02-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structural Determinants of Substrate Recognition in the HAD Superfamily Member d-glycero-d-manno-Heptose-1,7-bisphosphate Phosphatase (GmhB) .
Biochemistry, 49, 2010
|
|
3L8H
 
 | | Crystal Structure of D,D-heptose 1.7-bisphosphate phosphatase from B. bronchiseptica complexed with magnesium and phosphate | | Descriptor: | FORMIC ACID, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Nguyen, H, Peisach, E, Allen, K.N. | | Deposit date: | 2009-12-31 | | Release date: | 2010-02-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural Determinants of Substrate Recognition in the HAD Superfamily Member d-glycero-d-manno-Heptose-1,7-bisphosphate Phosphatase (GmhB) .
Biochemistry, 49, 2010
|
|
5VGX
 
 | |
5VGV
 
 | |
5TTK
 
 | |
5WHL
 
 | |
5WIY
 
 | | Kelch domain of human Keap1 bound to small molecule inhibitor fragment: 4-amino-1,7-dihydro-6H-pyrazolo[3,4-d]pyrimidine-6-thione | | Descriptor: | 4-amino-1,7-dihydro-6H-pyrazolo[3,4-d]pyrimidine-6-thione, Kelch-like ECH-associated protein 1, SULFATE ION | | Authors: | Carolan, J.P, Lynch, A.J, Allen, K.N. | | Deposit date: | 2017-07-20 | | Release date: | 2018-09-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Interaction Energetics and Druggability of the Protein-Protein Interaction between Kelch-like ECH-Associated Protein 1 (KEAP1) and Nuclear Factor Erythroid 2 Like 2 (Nrf2).
Biochemistry, 59, 2020
|
|
2RAV
 
 | |
2RBK
 
 | |
2RB5
 
 | |
2RAR
 
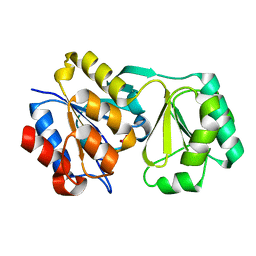 | |
4NEJ
 
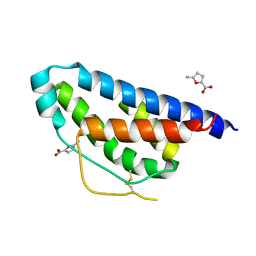 | | Small molecular fragment bound to crystal contact interface of Interleukin-2 | | Descriptor: | 5-methylfuran-2-carboxylic acid, Interleukin-2 | | Authors: | Brenke, R, Jehle, S, Vajda, S, Allen, K.N, Kozakov, D. | | Deposit date: | 2013-10-29 | | Release date: | 2014-11-19 | | Method: | X-RAY DIFFRACTION (1.919 Å) | | Cite: | Small molecular fragments bound to binding energy hot-spot in crystal contact interface of Interleukin-2
To be Published
|
|
4NEM
 
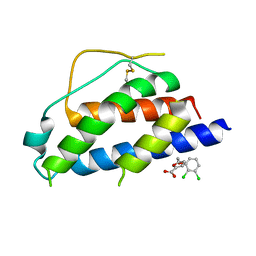 | | Small molecular fragment bound to crystal contact interface of Interleukin-2 | | Descriptor: | 5-[(2,3-dichlorophenoxy)methyl]furan-2-carboxylic acid, Interleukin-2 | | Authors: | Jehle, S, Brenke, R, Vajda, S, Allen, K.N, Kozakov, D. | | Deposit date: | 2013-10-29 | | Release date: | 2014-11-19 | | Method: | X-RAY DIFFRACTION (1.934 Å) | | Cite: | Small molecular fragments bound to binding energy hot-spot in crystal contact interface of Interleukin-2
To be Published
|
|
4QD9
 
 | |
4OFZ
 
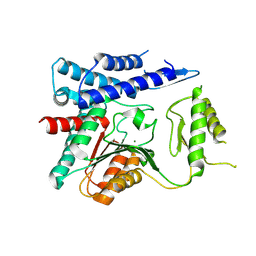 | | Structure of unliganded trehalose-6-phosphate phosphatase from Brugia malayi | | Descriptor: | MAGNESIUM ION, Trehalose-phosphatase | | Authors: | Farelli, J.D, Allen, K.N, Carlow, C.K.S, Dunaway-Mariano, D. | | Deposit date: | 2014-01-15 | | Release date: | 2014-07-16 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the Trehalose-6-phosphate Phosphatase from Brugia malayi Reveals Key Design Principles for Anthelmintic Drugs.
Plos Pathog., 10, 2014
|
|
4QDA
 
 | |
4QD7
 
 | |
4QDB
 
 | |
4QD8
 
 | |
8E37
 
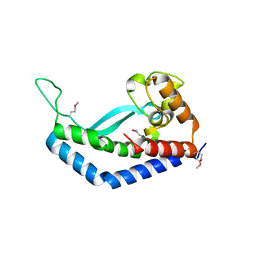 | | Structure of Campylobacter concisus wild-type SeMet PglC | | Descriptor: | N,N'-diacetylbacilliosaminyl-1-phosphate transferase | | Authors: | Vuksanovic, N, Ray, L.C, Imperiali, B, Allen, K.N. | | Deposit date: | 2022-08-16 | | Release date: | 2023-09-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Synergistic computational and experimental studies of a phosphoglycosyl transferase membrane/ligand ensemble.
J.Biol.Chem., 299, 2023
|
|
8DVW
 
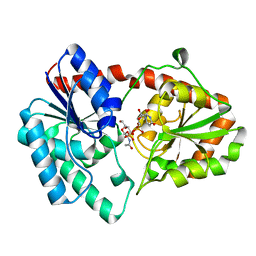 | | Structure of the Campylobacter concisus glycosyltransferase PglA R203Q | | Descriptor: | N, N'-diacetylbacillosaminyl-diphospho-undecaprenol alpha-1,3-N-acetylgalactosaminyltransferase, URIDINE-DIPHOSPHATE-N-ACETYLGALACTOSAMINE | | Authors: | Vuksanovic, N, Clasman, J.R, Bernstein, H.M, Imperiali, B, Allen, K.N. | | Deposit date: | 2022-07-30 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Specificity determinants revealed by the structure of glycosyltransferase Campylobacter concisus PglA.
Protein Sci., 33, 2024
|
|
8DVZ
 
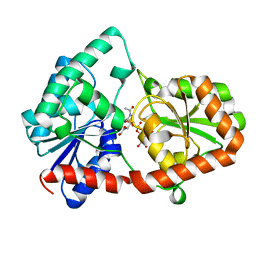 | | Structure of the Campylobacter concisus glycosyltransferase PglA R282V variant | | Descriptor: | N, N'-diacetylbacillosaminyl-diphospho-undecaprenol alpha-1,3-N-acetylgalactosaminyltransferase, URIDINE-DIPHOSPHATE-N-ACETYLGALACTOSAMINE | | Authors: | Vuksanovic, N, Clasman, J.R, Bernstein, H.M, Imperiali, B, Allen, K.N. | | Deposit date: | 2022-07-30 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Specificity determinants revealed by the structure of glycosyltransferase Campylobacter concisus PglA.
Protein Sci., 33, 2024
|
|
8DQD
 
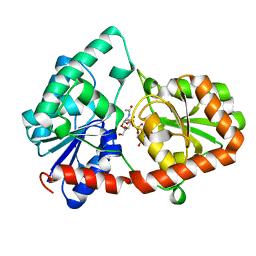 | | Structure of the Campylobacter concisus glycosyltransferase PglA | | Descriptor: | N, N'-diacetylbacillosaminyl-diphospho-undecaprenol alpha-1,3-N-acetylgalactosaminyltransferase, URIDINE-DIPHOSPHATE-N-ACETYLGALACTOSAMINE | | Authors: | Vuksanovic, N, Clasman, J.R, Bernstein, H.M, Imperiali, B, Allen, K.N. | | Deposit date: | 2022-07-18 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Specificity determinants revealed by the structure of glycosyltransferase Campylobacter concisus PglA.
Protein Sci., 33, 2024
|
|
8EEG
 
 | |
8EEM
 
 | |
