6L3H
 
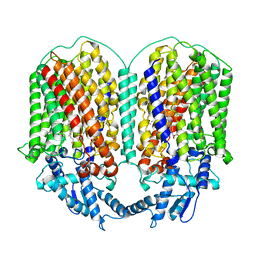 | | Cryo-EM structure of dimeric quinol dependent Nitric Oxide Reductase (qNOR) from the pathogen Neisseria meninigitidis | | Descriptor: | CALCIUM ION, FE (III) ION, Nitric-oxide reductase, ... | | Authors: | Jamali, M.M.A, Gopalasingam, C.C, Johnson, R.M, Tosha, T, Muench, S.P, Muramoto, K, Antonyuk, S.V, Shiro, Y, Hasnain, S.S. | | Deposit date: | 2019-10-11 | | Release date: | 2020-04-01 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | The active form of quinol-dependent nitric oxide reductase fromNeisseria meningitidisis a dimer.
Iucrj, 7, 2020
|
|
6L1X
 
 | | Quinol-dependent nitric oxide reductase (qNOR) from Neisseria meningitidis in the monomeric oxidized state with zinc complex. | | Descriptor: | CALCIUM ION, FE (III) ION, Nitric-oxide reductase, ... | | Authors: | Jamali, M.M.A, Antonyuk, S.V, Tosha, T, Muramoto, K, Hasnain, S.S, Shiro, Y. | | Deposit date: | 2019-10-01 | | Release date: | 2020-04-01 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | The active form of quinol-dependent nitric oxide reductase fromNeisseria meningitidisis a dimer.
Iucrj, 7, 2020
|
|
1DU5
 
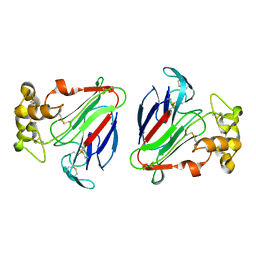 | | THE CRYSTAL STRUCTURE OF ZEAMATIN. | | Descriptor: | ZEAMATIN | | Authors: | Batalia, M.A, Monzingo, A.F, Ernst, S, Roberts, W, Robertus, J.D. | | Deposit date: | 2000-01-14 | | Release date: | 2000-02-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of the antifungal protein zeamatin, a member of the thaumatin-like, PR-5 protein family.
Nat.Struct.Biol., 3, 1996
|
|
6T2E
 
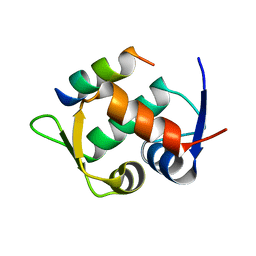 | | Multicomponent Peptide Stapling as a Diversity-Driven Tool for the Development of Inhibitors of Protein-Protein Interactions | | Descriptor: | E3 ubiquitin-protein ligase Mdm2, Stapled peptide GAR300-Gm | | Authors: | Groves, R.M, Ali, M.A, Atmaj, J, van Oosterwijk, N, Domling, A, Rivera, G.D, Ricardo, G.M. | | Deposit date: | 2019-10-08 | | Release date: | 2020-01-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Multicomponent Peptide Stapling as a Diversity-Driven Tool for the Development of Inhibitors of Protein-Protein Interactions.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6T2F
 
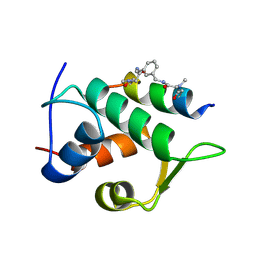 | | Multicomponent Peptide Stapling as a Diversity-Driven Tool for the Development of Inhibitors of Protein-Protein Interactions | | Descriptor: | 2-(methylamino)-~{N}-[[3-[[2-(methylamino)ethanoylamino]methyl]phenyl]methyl]ethanamide, E3 ubiquitin-protein ligase Mdm2, MDM2 in complex with GAR300-Am | | Authors: | Groves, R.M, Ali, M.A, Atmaj, J, van Oosterwijk, N, Domling, A, Rivera, G.D, Ricardo, G.M. | | Deposit date: | 2019-10-08 | | Release date: | 2020-01-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Multicomponent Peptide Stapling as a Diversity-Driven Tool for the Development of Inhibitors of Protein-Protein Interactions.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6T2D
 
 | | Multicomponent Peptide Stapling as a Diversity-Driven Tool for the Development of Inhibitors of Protein-Protein Interactions | | Descriptor: | 2-(methylamino)-~{N}-[[4-[[2-(methylamino)ethanoylamino]methyl]phenyl]methyl]ethanamide, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DIMETHYL SULFOXIDE, ... | | Authors: | Groves, R.M, Ali, M.A, Atmaj, J, van Oosterwijk, N, Domling, A, Rivera, G.D, Ricardo, G.M. | | Deposit date: | 2019-10-08 | | Release date: | 2020-01-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Multicomponent Peptide Stapling as a Diversity-Driven Tool for the Development of Inhibitors of Protein-Protein Interactions.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
1QR1
 
 | | POOR BINDING OF A HER-2/NEU EPITOPE (GP2) TO HLA-A2.1 IS DUE TO A LACK OF INTERACTIONS IN THE CENTER OF THE PEPTIDE | | Descriptor: | BETA-2 MICROGLOBULIN, GP2 PEPTIDE, HLA-A2.1 HEAVY CHAIN | | Authors: | Kuhns, J.J, Batalia, M.A, Yan, S, Collins, E.J. | | Deposit date: | 1999-06-17 | | Release date: | 2000-01-01 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Poor binding of a HER-2/neu epitope (GP2) to HLA-A2.1 is due to a lack of interactions with the center of the peptide.
J.Biol.Chem., 274, 1999
|
|
9GDM
 
 | |
8TV4
 
 | | NMR structure of temporin L in solution | | Descriptor: | Temporin-1Tl peptide | | Authors: | McShan, A.C, Jia, R, Halim, M.A. | | Deposit date: | 2023-08-17 | | Release date: | 2023-09-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Antiviral peptides inhibiting the main protease of SARS-CoV-2 investigated by computational screening and in vitro protease assay.
J.Pept.Sci., 30, 2024
|
|
2MBS
 
 | | NMR solution structure of oxidized KpDsbA | | Descriptor: | Thiol:disulfide interchange protein | | Authors: | Kurth, F, Rimmer, K, Premkumar, L, Mohanty, B, Duprez, W, Halili, M.A, Shouldice, S.R, Heras, B, Fairlie, D.P, Scanlon, M.J, Martin, J.L. | | Deposit date: | 2013-08-03 | | Release date: | 2013-12-11 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Comparative Sequence, Structure and Redox Analyses of Klebsiella pneumoniae DsbA Show That Anti-Virulence Target DsbA Enzymes Fall into Distinct Classes.
Plos One, 8, 2013
|
|
