5AGY
 
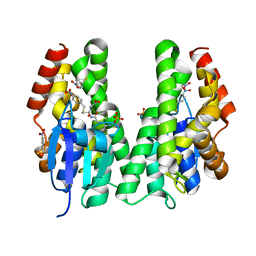 | | CRYSTAL STRUCTURE OF A TAU CLASS GST MUTANT FROM GLYCINE | | Descriptor: | 4-NITROPHENYL METHANETHIOL, GLUTATHIONE S-TRANSFERASE, PHOSPHATE ION, ... | | Authors: | Axarli, I, Muleta, A.W, Vlachakis, D, Kossida, S, Kotzia, G, Dhavala, P, Papageorgiou, A.C, Labrou, N.E. | | Deposit date: | 2015-02-04 | | Release date: | 2015-12-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Directed Evolution of Tau Class Glutathione Transferases Reveals a Site that Regulates Catalytic Efficiency and Masks Cooperativity.
Biochem.J., 473, 2016
|
|
2F98
 
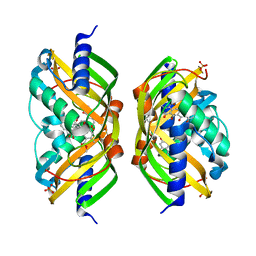 | | Crystal structure of the polyketide cyclase AknH with bound substrate and product analogue: implications for catalytic mechanism and product stereoselectivity. | | Descriptor: | Aklanonic Acid methyl Ester Cyclase, AknH, METHYL 5,7-DIHYDROXY-2-METHYL-4,6,11-TRIOXO-3,4,6,11-TETRAHYDROTETRACENE-1-CARBOXYLATE, ... | | Authors: | Kallio, P, Sultana, A, Neimi, J, Mantsala, P, Schneider, G. | | Deposit date: | 2005-12-05 | | Release date: | 2006-02-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the polyketide cyclase AknH with bound substrate and product analogue: implications for catalytic mechanism and product stereoselectivity.
J.Mol.Biol., 357, 2006
|
|
2F99
 
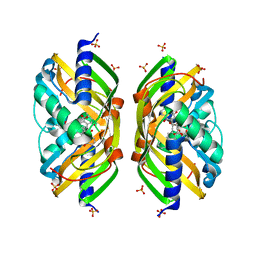 | | Crystal structure of the polyketide cyclase AknH with bound substrate and product analogue: implications for catalytic mechanism and product stereoselectivity. | | Descriptor: | Aklanonic Acid methyl Ester Cyclase, AknH, SULFATE ION, ... | | Authors: | Kallio, P, Sultana, A, Neimi, J, Mantsala, P, Schneider, G. | | Deposit date: | 2005-12-05 | | Release date: | 2006-02-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the polyketide cyclase AknH with bound substrate and product analogue: implications for catalytic mechanism and product stereoselectivity.
J.Mol.Biol., 357, 2006
|
|
1QRD
 
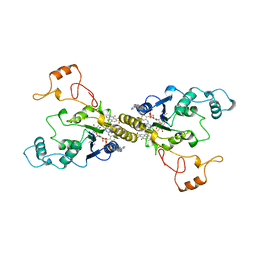 | | QUINONE REDUCTASE/FAD/CIBACRON BLUE/DUROQUINONE COMPLEX | | Descriptor: | CIBACRON BLUE, DUROQUINONE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Li, R, Bianchet, M.A, Talalay, P, Amzel, L.M. | | Deposit date: | 1995-07-28 | | Release date: | 1996-10-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The three-dimensional structure of NAD(P)H:quinone reductase, a flavoprotein involved in cancer chemoprotection and chemotherapy: mechanism of the two-electron reduction.
Proc.Natl.Acad.Sci.USA, 92, 1995
|
|
6FM2
 
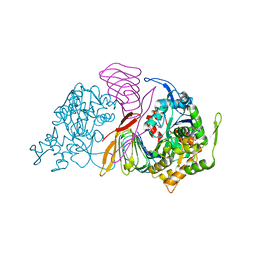 | | CARP domain of mouse cyclase-associated protein 1 (CAP1) bound to ADP-actin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Kotila, T.M, Kogan, K, Lappalainen, P. | | Deposit date: | 2018-01-30 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of actin monomer re-charging by cyclase-associated protein.
Nat Commun, 9, 2018
|
|
1O0R
 
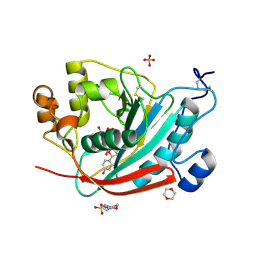 | | Crystal structure of the catalytic domain of bovine beta1,4-galactosyltransferase complex with UDP-galactose | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GALACTOSE-URIDINE-5'-DIPHOSPHATE, ... | | Authors: | Ramakrishnan, B, Balaji, P.V, Qasba, P.K. | | Deposit date: | 2003-02-24 | | Release date: | 2003-03-04 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | CRYSTAL STRUCTURE OF BETA 1,4-GALACTOSYLTRANSFERASE COMPLEX WITH UDP-GAL
REVEALS AN OLIGOSACCHARIDE ACCEPTOR BINDING SITE
J.Mol.Biol., 318, 2002
|
|
4AYA
 
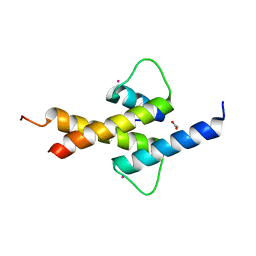 | | Crystal structure of ID2 HLH homodimer at 2.1A resolution | | Descriptor: | ACETATE ION, DNA-BINDING PROTEIN INHIBITOR ID-2, POTASSIUM ION | | Authors: | Wong, M.V, Jiang, S, Palasingam, P, Kolatkar, P.R. | | Deposit date: | 2012-06-19 | | Release date: | 2012-11-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.103 Å) | | Cite: | A Divalent Ion is Crucial in the Structure and Dominant-Negative Function of Id Proteins, a Class of Helix-Loop-Helix Transcription Regulators.
Plos One, 7, 2012
|
|
5GIB
 
 | | Succinic acid bound trypsin crystallized as dimer | | Descriptor: | CALCIUM ION, Cationic trypsin, SUCCINIC ACID | | Authors: | Kutumbarao, N.H.V, Manohar, R, KarthiK, L, Malathy, P, Gunasekaran, K, Velmurugan, D. | | Deposit date: | 2016-06-22 | | Release date: | 2016-08-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.697 Å) | | Cite: | Trypsin bound with Succinic acid
To Be Published
|
|
1QR2
 
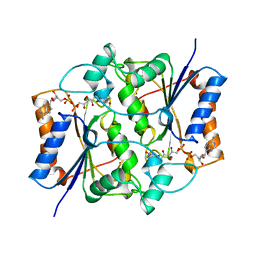 | | HUMAN QUINONE REDUCTASE TYPE 2 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, PROTEIN (QUINONE REDUCTASE TYPE 2), ZINC ION | | Authors: | Foster, C, Bianchet, M.A, Talalay, P, Amzel, L.M. | | Deposit date: | 1999-04-15 | | Release date: | 1999-08-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of human quinone reductase type 2, a metalloflavoprotein.
Biochemistry, 38, 1999
|
|
1BSJ
 
 | | COBALT DEFORMYLASE INHIBITOR COMPLEX FROM E.COLI | | Descriptor: | (S)-2-(PHOSPHONOXY)CAPROYL-L-LEUCYL-P-NITROANILIDE, COBALT (II) ION, PHOSPHATE ION, ... | | Authors: | Hao, B, Gong, W, Rajagopalan, P.T, Hu, Y, Pei, D, Chan, M.K. | | Deposit date: | 1998-08-28 | | Release date: | 2000-04-15 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for the design of antibiotics targeting peptide deformylase.
Biochemistry, 38, 1999
|
|
1BSK
 
 | | ZINC DEFORMYLASE INHIBITOR COMPLEX FROM E.COLI | | Descriptor: | (S)-2-(PHOSPHONOXY)CAPROYL-L-LEUCYL-P-NITROANILIDE, PHOSPHATE ION, PROTEIN (PEPTIDE DEFORMYLASE), ... | | Authors: | Hao, B, Gong, W, Rajagopalan, P.T, Hu, Y, Pei, D, Chan, M.K. | | Deposit date: | 1998-08-28 | | Release date: | 2000-04-15 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for the design of antibiotics targeting peptide deformylase.
Biochemistry, 38, 1999
|
|
4EX6
 
 | | Crystal structure of the alnumycin P phosphatase AlnB | | Descriptor: | AlnB, BORIC ACID, MAGNESIUM ION | | Authors: | Oja, T, Niiranen, L, Sandalova, T, Klika, K.D, Niemi, J, Mantsala, P, Schneider, G, Metsa-Ketela, M. | | Deposit date: | 2012-04-30 | | Release date: | 2013-01-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural basis for C-ribosylation in the alnumycin A biosynthetic pathway.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4E6S
 
 | | Crystal structure of the SCAN domain from mouse Zfp206 | | Descriptor: | Zinc finger and SCAN domain-containing protein 10 | | Authors: | Liang, Y, Choo, S.H, Rossbach, M, Baburajendran, N, Palasingam, P, Kolatkar, P.R. | | Deposit date: | 2012-03-15 | | Release date: | 2012-05-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal optimization and preliminary diffraction data analysis of the SCAN domain of Zfp206.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
1QPV
 
 | | YEAST COFILIN | | Descriptor: | YEAST COFILIN | | Authors: | Fedorov, A.A, Lappalainen, P, Fedorov, E.V, Drubin, D.G, Almo, S.C. | | Deposit date: | 1999-05-29 | | Release date: | 1999-06-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure determination of yeast cofilin.
Nat.Struct.Biol., 4, 1997
|
|
1QD9
 
 | | Bacillus subtilis YABJ | | Descriptor: | ACETIC ACID, ETHYL MERCURY ION, MERCURY (II) ION, ... | | Authors: | Smith, J.L, Sinha, S, Rappu, P, Lange, S.C, Mantsala, P, Zalkin, H. | | Deposit date: | 1999-07-09 | | Release date: | 1999-11-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of Bacillus subtilis YabJ, a purine regulatory protein and member of the highly conserved YjgF family.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
2H1P
 
 | | THE THREE-DIMENSIONAL STRUCTURES OF A POLYSACCHARIDE BINDING ANTIBODY TO CRYPTOCOCCUS NEOFORMANS AND ITS COMPLEX WITH A PEPTIDE FROM A PHAGE DISPLAY LIBRARY: IMPLICATIONS FOR THE IDENTIFICATION OF PEPTIDE MIMOTOPES | | Descriptor: | 2H1, PA1 | | Authors: | Young, A.C.M, Valadon, P, Casadevall, A, Scharff, M.D, Sacchettini, J.C. | | Deposit date: | 1997-11-12 | | Release date: | 1998-01-28 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The three-dimensional structures of a polysaccharide binding antibody to Cryptococcus neoformans and its complex with a peptide from a phage display library: implications for the identification of peptide mimotopes.
J.Mol.Biol., 274, 1997
|
|
1SJW
 
 | | Structure of polyketide cyclase SnoaL | | Descriptor: | METHYL 5,7-DIHYDROXY-2-METHYL-4,6,11-TRIOXO-3,4,6,11-TETRAHYDROTETRACENE-1-CARBOXYLATE, nogalonic acid methyl ester cyclase | | Authors: | Sultana, A, Kallio, P, Jansson, A, Wang, J.S, Neimi, J, Mantsala, P, Schneider, G, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2004-03-04 | | Release date: | 2004-04-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure of the polyketide cyclase SnoaL reveals a novel mechanism for enzymatic aldol condensation.
Embo J., 23, 2004
|
|
1CYX
 
 | | QUINOL OXIDASE (PERIPLASMIC FRAGMENT OF SUBUNIT II WITH ENGINEERED CU-A BINDING SITE)(CYOA) | | Descriptor: | CYOA, DINUCLEAR COPPER ION | | Authors: | Wilmanns, M, Lappalainen, P, Kelly, M, Sauer-Eriksson, E, Saraste, M. | | Deposit date: | 1995-08-22 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the membrane-exposed domain from a respiratory quinol oxidase complex with an engineered dinuclear copper center.
Proc.Natl.Acad.Sci.USA, 92, 1995
|
|
3WR1
 
 | | Crystal structure of Cormorant (Phalacrocorax carbo) hemoglobin | | Descriptor: | Hemoglobin subunit alpha-A, Hemoglobin subunit beta, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Jagadeesan, G, Vinodh Kumar, V, Peters, H.G, Malathy, P, Harikrishna Etti, S, Gunasekaran, K, Aravindhan, S. | | Deposit date: | 2014-02-09 | | Release date: | 2014-03-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structure of Cormorant (Phalacrocorax carbo) hemoglobin
To be Published
|
|
1DFF
 
 | | PEPTIDE DEFORMYLASE | | Descriptor: | PEPTIDE DEFORMYLASE, ZINC ION | | Authors: | Chan, M.K, Gong, W, Rajagopalan, P.T.R, Hao, B, Tsai, C.M, Pei, D. | | Deposit date: | 1997-08-19 | | Release date: | 1998-09-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Crystal structure of the Escherichia coli peptide deformylase.
Biochemistry, 36, 1997
|
|
2INX
 
 | | Crystal Structure of Ketosteroid Isomerase D40N from Pseudomonas putida (pKSI) with bound 2,6-difluorophenol | | Descriptor: | 2,6-DIFLUOROPHENOL, Steroid delta-isomerase | | Authors: | Martinez Caaveiro, J.M, Pybus, B, Ringe, D, Petsko, G.A, Sigala, P, Kraut, D, Herschlag, D. | | Deposit date: | 2006-10-09 | | Release date: | 2007-10-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Testing geometrical discrimination within an enzyme active site: constrained hydrogen bonding in the ketosteroid isomerase oxyanion hole.
J.Am.Chem.Soc., 130, 2008
|
|
1JY6
 
 | |
1JY4
 
 | |
1BS9
 
 | | ACETYLXYLAN ESTERASE FROM P. PURPUROGENUM REFINED AT 1.10 ANGSTROMS | | Descriptor: | ACETYL XYLAN ESTERASE, SULFATE ION | | Authors: | Ghosh, D, Erman, M, Sawicki, M.W, Lala, P, Weeks, D.R, Li, N, Pangborn, W, Thiel, D.J, Jornvall, H, Eyzaguirre, J. | | Deposit date: | 1998-09-01 | | Release date: | 1999-05-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Determination of a protein structure by iodination: the structure of iodinated acetylxylan esterase.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
4KWI
 
 | | The crystal structure of angucycline C-6 ketoreductase LanV with bound NADP and 11-deoxy-6-oxylandomycinone | | Descriptor: | 1,8-dihydroxy-3-methyltetraphene-6,7,12(5H)-trione, DI(HYDROXYETHYL)ETHER, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Paananen, P, Patrikainen, P, Mantsala, P, Niemi, J, Niiranen, L, Metsa-Ketela, M. | | Deposit date: | 2013-05-24 | | Release date: | 2013-07-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and functional analysis of angucycline C-6 ketoreductase LanV involved in landomycin biosynthesis.
Biochemistry, 52, 2013
|
|
