7X30
 
 | | Capsid structure of Staphylococcus jumbo bacteriophage S6 | | Descriptor: | Hoc-like protein ORF90, Major structural protein ORF12 | | Authors: | Koibuchi, W, Uchiyama, J, Matsuzaki, S, Murata, K, Iwasaki, K, Miyazaki, N. | | Deposit date: | 2022-02-27 | | Release date: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Capsid structure of Staphylococcus jumbo bacteriophage S6
To Be Published
|
|
7DOU
 
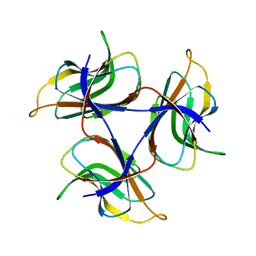 | | Trimeric cement protein structure of Helicobacter pylori bacteriophage KHP40 | | Descriptor: | Cement protein gp16 | | Authors: | Kamiya, R, Uchiyama, J, Matsuzaki, S, Murata, K, Iwasaki, K, Miyazaki, N. | | Deposit date: | 2020-12-17 | | Release date: | 2021-10-27 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Acid-stable capsid structure of Helicobacter pylori bacteriophage KHP30 by single-particle cryoelectron microscopy.
Structure, 30, 2022
|
|
5AWW
 
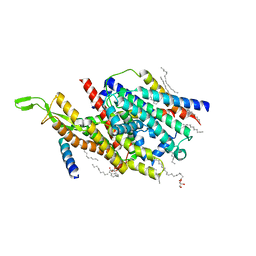 | | Precise Resting State of Thermus thermophilus SecYEG | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Protein translocase subunit SecE, Protein translocase subunit SecY, ... | | Authors: | Tanaka, Y, Sugano, Y, Takemoto, M, Kusakizako, T, Kumazaki, K, Ishitani, R, Nureki, O, Tsukazaki, T. | | Deposit date: | 2015-07-10 | | Release date: | 2015-11-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.724 Å) | | Cite: | Crystal Structures of SecYEG in Lipidic Cubic Phase Elucidate a Precise Resting and a Peptide-Bound State.
Cell Rep, 13, 2015
|
|
2O7C
 
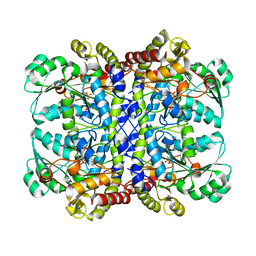 | | Crystal structure of L-methionine-lyase from Pseudomonas | | Descriptor: | Methionine gamma-lyase, SULFATE ION | | Authors: | Misaki, S, Takimoto, A, Takakura, T, Yoshioka, T, Yamashita, M, Tamura, T, Tanaka, H, Inagaki, K. | | Deposit date: | 2006-12-10 | | Release date: | 2007-12-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the antitumour enzyme L-methionine gamma-lyase from Pseudomonas putida at 1.8 A resolution
J.Biochem.(Tokyo), 141, 2007
|
|
7DOD
 
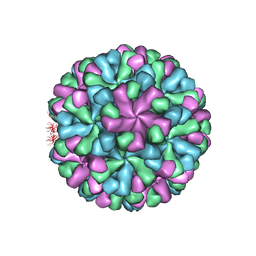 | | Capsid structure of human sapovirus | | Descriptor: | Calicivirin | | Authors: | Miyazaki, N, Murakami, K, Oka, T, Iwasaki, K, Katayama, K, Murata, K. | | Deposit date: | 2020-12-14 | | Release date: | 2021-12-15 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Atomic structure of human sapovirus capsid by single particle cryo-electron microscopy
To Be Published
|
|
6IUK
 
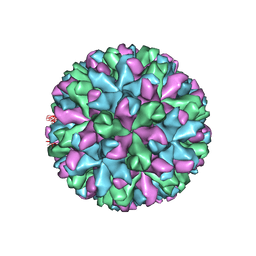 | | Cryo-EM structure of Murine Norovirus capsid | | Descriptor: | Major capsid protein VP1 | | Authors: | Song, C, Miyazaki, N, Iwasaki, K, Katayama, K, Murata, K. | | Deposit date: | 2018-11-28 | | Release date: | 2020-02-26 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Dynamic rotation of the protruding domain enhances the infectivity of norovirus.
Plos Pathog., 16, 2020
|
|
1AYG
 
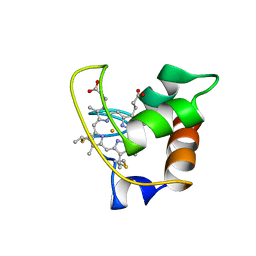 | | SOLUTION STRUCTURE OF CYTOCHROME C-552, NMR, 20 STRUCTURES | | Descriptor: | CYTOCHROME C-552, HEME C | | Authors: | Hasegawa, J, Yoshida, T, Yamazaki, T, Sambongi, Y, Yu, Y, Igarashi, Y, Kodama, T, Yamazaki, K, Hakusui, H, Kyogoku, Y, Kobayashi, Y. | | Deposit date: | 1997-11-04 | | Release date: | 1998-11-25 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of thermostable cytochrome c-552 from Hydrogenobacter thermophilus determined by 1H-NMR spectroscopy.
Biochemistry, 37, 1998
|
|
3MPW
 
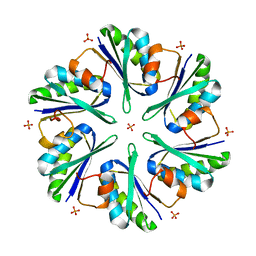 | | Structure of EUTM in 2-D protein membrane | | Descriptor: | Ethanolamine utilization protein eutM, PHOSPHATE ION | | Authors: | Sagermann, M, Takenoya, M, Nikolakakis, K. | | Deposit date: | 2010-04-27 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystallographic insights into the pore structures and mechanisms of the EutL and EutM shell proteins of the ethanolamine-utilizing microcompartment of Escherichia coli.
J.Bacteriol., 192, 2010
|
|
1C9F
 
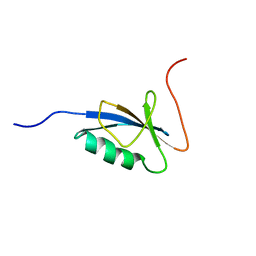 | |
6KNC
 
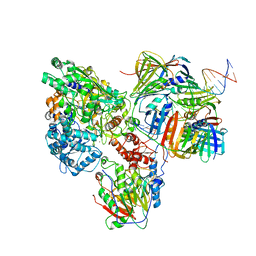 | | PolD-PCNA-DNA (form B) | | Descriptor: | DNA polymerase D DP2 (DNA polymerase II large) subunit, DNA polymerase II small subunit, DNA polymerase sliding clamp 1, ... | | Authors: | Mayanagi, K, Oki, K, Miyazaki, N, Ishino, S, Yamagami, T, Iwasaki, K, Kohda, D, Morikawa, K, Shirai, T, Ishino, Y. | | Deposit date: | 2019-08-05 | | Release date: | 2020-08-05 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (9.3 Å) | | Cite: | Two conformations of DNA polymerase D-PCNA-DNA, an archaeal replisome complex, revealed by cryo-electron microscopy.
Bmc Biol., 18, 2020
|
|
6KNB
 
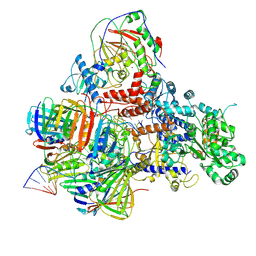 | | PolD-PCNA-DNA (form A) | | Descriptor: | DNA polymerase D DP2 (DNA polymerase II large) subunit, DNA polymerase II small subunit, DNA polymerase sliding clamp 1, ... | | Authors: | Mayanagi, K, Oki, K, Miyazaki, N, Ishino, S, Yamagami, T, Iwasaki, K, Kohda, D, Morikawa, K, Shirai, T, Ishino, Y. | | Deposit date: | 2019-08-05 | | Release date: | 2020-08-05 | | Last modified: | 2021-02-17 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Two conformations of DNA polymerase D-PCNA-DNA, an archaeal replisome complex, revealed by cryo-electron microscopy.
Bmc Biol., 18, 2020
|
|
5CH4
 
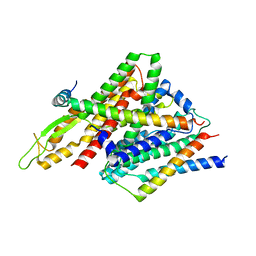 | | Peptide-Bound State of Thermus thermophilus SecYEG | | Descriptor: | Protein translocase subunit SecE, Protein translocase subunit SecY, Putative preprotein translocase, ... | | Authors: | Tanaka, Y, Sugano, Y, Takemoto, M, Kusakizako, T, Kumazaki, K, Ishitani, R, Nureki, O, Tsukazaki, T. | | Deposit date: | 2015-07-10 | | Release date: | 2015-11-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.64 Å) | | Cite: | Crystal Structures of SecYEG in Lipidic Cubic Phase Elucidate a Precise Resting and a Peptide-Bound State.
Cell Rep, 13, 2015
|
|
3MPY
 
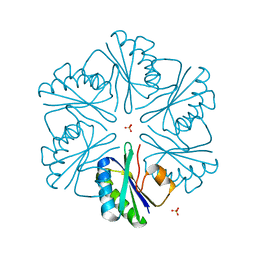 | | Structure of EUTM in 2-D protein membrane | | Descriptor: | Ethanolamine utilization protein eutM, SULFATE ION | | Authors: | Sagermann, M, Takenoya, M, Nikolakakis, K. | | Deposit date: | 2010-04-27 | | Release date: | 2010-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic insights into the pore structures and mechanisms of the EutL and EutM shell proteins of the ethanolamine-utilizing microcompartment of Escherichia coli.
J.Bacteriol., 192, 2010
|
|
3GFH
 
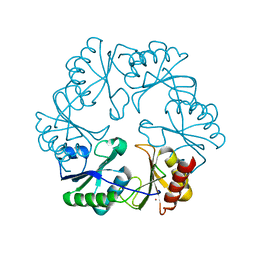 | |
3MPV
 
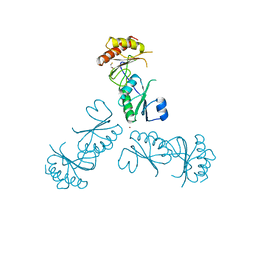 | | Structure of EUTL in the zinc-induced open form | | Descriptor: | BETA-MERCAPTOETHANOL, Ethanolamine utilization protein eutL, ZINC ION | | Authors: | Sagermann, M, Takenoya, M, Nikolakakis, K. | | Deposit date: | 2010-04-27 | | Release date: | 2010-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystallographic insights into the pore structures and mechanisms of the EutL and EutM shell proteins of the ethanolamine-utilizing microcompartment of Escherichia coli.
J.Bacteriol., 192, 2010
|
|
1A05
 
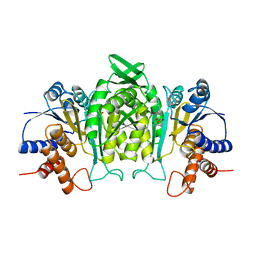 | | CRYSTAL STRUCTURE OF THE COMPLEX OF 3-ISOPROPYLMALATE DEHYDROGENASE FROM THIOBACILLUS FERROOXIDANS WITH 3-ISOPROPYLMALATE | | Descriptor: | 3-ISOPROPYLMALATE DEHYDROGENASE, 3-ISOPROPYLMALIC ACID, MAGNESIUM ION | | Authors: | Imada, K, Inagaki, K, Matsunami, H, Kawaguchi, H, Tanaka, H, Tanaka, N, Namba, K. | | Deposit date: | 1997-12-09 | | Release date: | 1998-06-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of 3-isopropylmalate dehydrogenase in complex with 3-isopropylmalate at 2.0 A resolution: the role of Glu88 in the unique substrate-recognition mechanism.
Structure, 6, 1998
|
|
8WDV
 
 | | Photosynthetic LH1-RC complex from the purple sulfur bacterium Allochromatium vinosum purified by Ca2+-DEAE | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (2S)-3-hydroxypropane-1,2-diyl dihexadecanoate, Antenna complex alpha/beta subunit, ... | | Authors: | Tani, K, Kanno, R, Harada, A, Kobayashi, A, Minamino, A, Nakamura, N, Ji, X.-C, Purba, E.R, Hall, M, Yu, L.-J, Madigan, M.T, Mizoguchi, A, Iwasaki, K, Humbel, B.M, Kimura, Y, Wang-Otomo, Z.-Y. | | Deposit date: | 2023-09-16 | | Release date: | 2024-02-21 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.24 Å) | | Cite: | High-resolution structure and biochemical properties of the LH1-RC photocomplex from the model purple sulfur bacterium, Allochromatium vinosum.
Commun Biol, 7, 2024
|
|
5GJE
 
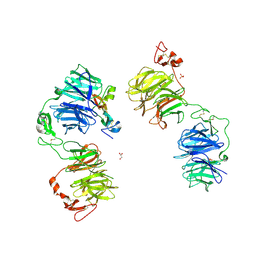 | | Three-dimensional reconstruction of human LRP6 ectodomain complexed with Dkk1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Matoba, K, Mihara, E, Tamura-Kawakami, K, Hirai, H, Thompson, S, Iwasaki, K, Takagi, J. | | Deposit date: | 2016-06-29 | | Release date: | 2017-01-18 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (21 Å) | | Cite: | Conformational Freedom of the LRP6 Ectodomain Is Regulated by N-glycosylation and the Binding of the Wnt Antagonist Dkk1
Cell Rep, 18, 2017
|
|
8WDU
 
 | | Photosynthetic LH1-RC complex from the purple sulfur bacterium Allochromatium vinosum purified by sucrose density | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (2S)-3-hydroxypropane-1,2-diyl dihexadecanoate, Antenna complex alpha/beta subunit, ... | | Authors: | Tani, K, Kanno, R, Harada, A, Kobayashi, A, Minamino, A, Nakamura, N, Ji, X.-C, Purba, E.R, Hall, M, Yu, L.-J, Madigan, M.T, Mizoguchi, A, Iwasaki, K, Humbel, B.M, Kimura, Y, Wang-Otomo, Z.-Y. | | Deposit date: | 2023-09-16 | | Release date: | 2024-02-21 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.24 Å) | | Cite: | High-resolution structure and biochemical properties of the LH1-RC photocomplex from the model purple sulfur bacterium, Allochromatium vinosum.
Commun Biol, 7, 2024
|
|
6LXV
 
 | | Cryo-EM structure of phosphoketolase from Bifidobacterium longum | | Descriptor: | CALCIUM ION, Phosphoketolase, THIAMINE DIPHOSPHATE | | Authors: | Nakata, K, Miyazaki, N, Yamaguchi, H, Hirose, M, Miyano, H, Mizukoshi, T, Kashiwagi, T, Iwasaki, K. | | Deposit date: | 2020-02-12 | | Release date: | 2021-02-17 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.1 Å) | | Cite: | High-resolution structure of phosphoketolase from Bifidobacterium longum determined by cryo-EM single-particle analysis.
J.Struct.Biol., 214, 2022
|
|
1UKJ
 
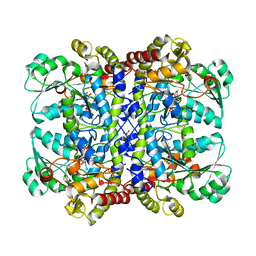 | | Detailed structure of L-Methionine-Lyase from Pseudomonas putida | | Descriptor: | Methionine gamma-lyase, SULFATE ION | | Authors: | Misaki, S, Takimoto, A, Takakura, T, Yoshioka, T, Yamashita, M, Tamura, T, Tanaka, H, Inagaki, K. | | Deposit date: | 2003-08-24 | | Release date: | 2004-10-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Detailed structure of L-Methionine -Lyase from Pseudomonas putida
To be Published
|
|
6AB6
 
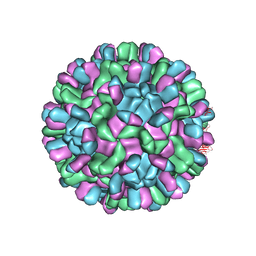 | | Cryo-EM structure of T=3 Penaeus vannamei nodavirus | | Descriptor: | CALCIUM ION, Capsid protein | | Authors: | Chen, N.C, Miyazaki, N, Yoshimura, M, Guan, H.H, Lin, C.C, Iwasaki, K, Chen, C.J. | | Deposit date: | 2018-07-20 | | Release date: | 2019-03-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The atomic structures of shrimp nodaviruses reveal new dimeric spike structures and particle polymorphism.
Commun Biol, 2, 2019
|
|
1RCH
 
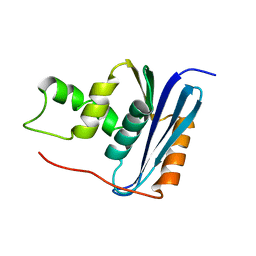 | | SOLUTION NMR STRUCTURE OF RIBONUCLEASE HI FROM ESCHERICHIA COLI, 8 STRUCTURES | | Descriptor: | RIBONUCLEASE HI | | Authors: | Yamazaki, T, Fujiwara, M, Kato, T, Yamasaki, K, Nagayama, K. | | Deposit date: | 1995-06-23 | | Release date: | 1997-02-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Ribonuclease Hi from Escherichia Coli
Biol.Pharm.Bull., 23, 2000
|
|
1RRB
 
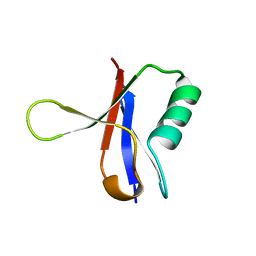 | | THE RAS-BINDING DOMAIN OF RAF-1 FROM RAT, NMR, 1 STRUCTURE | | Descriptor: | RAF PROTO-ONCOGENE SERINE/THREONINE-PROTEIN KINASE | | Authors: | Terada, T, Ito, Y, Shirouzu, M, Tateno, M, Hashimoto, K, Kigawa, T, Ebisuzaki, T, Takio, K, Shibata, T, Yokoyama, S, Smith, B.O, Laue, E.D, Cooper, J.A, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1998-03-26 | | Release date: | 1999-03-30 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance and molecular dynamics studies on the interactions of the Ras-binding domain of Raf-1 with wild-type and mutant Ras proteins.
J.Mol.Biol., 286, 1999
|
|
6AB5
 
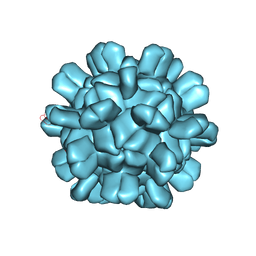 | | Cryo-EM structure of T=1 Penaeus vannamei nodavirus | | Descriptor: | Capsid protein | | Authors: | Chen, N.C, Miyazaki, N, Yoshimura, M, Guan, H.H, Lin, C.C, Iwasaki, K, Chen, C.J. | | Deposit date: | 2018-07-20 | | Release date: | 2019-03-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | The atomic structures of shrimp nodaviruses reveal new dimeric spike structures and particle polymorphism.
Commun Biol, 2, 2019
|
|
