1ZJB
 
 | | Crystal structure of the trehalulose synthase MutB from Pseudomonas mesoacidophila MX-45 (monoclinic form) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, Trehalulose synthase | | Authors: | Ravaud, S, Robert, X, Haser, R, Aghajari, N. | | Deposit date: | 2005-04-28 | | Release date: | 2006-10-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Expression, purification, crystallization and preliminary X-ray crystallographic studies of the trehalulose synthase MutB from Pseudomonas mesoacidophila MX-45.
Acta Crystallogr.,Sect.F, 61, 2005
|
|
1TVP
 
 | |
1TVN
 
 | |
1P6W
 
 | | Crystal structure of barley alpha-amylase isozyme 1 (AMY1) in complex with the substrate analogue, methyl 4I,4II,4III-tri-thiomaltotetraoside (thio-DP4) | | Descriptor: | CALCIUM ION, PROTEIN (Alpha-amylase type A isozyme), alpha-D-glucopyranose-(1-4)-4-thio-beta-D-glucopyranose, ... | | Authors: | Robert, X, Haser, R, Aghajari, N. | | Deposit date: | 2003-04-30 | | Release date: | 2003-10-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structure of barley alpha-amylase isozyme 1 reveals a novel role of domain C in substrate recognition and binding: a pair of sugar tongs
Structure, 11, 2003
|
|
3GBE
 
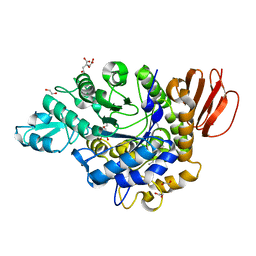 | | Crystal structure of the isomaltulose synthase SmuA from Protaminobacter rubrum in complex with the inhibitor deoxynojirimycin | | Descriptor: | 1,2-ETHANEDIOL, 1-DEOXYNOJIRIMYCIN, CITRATE ANION, ... | | Authors: | Ravaud, S, Robert, X, Haser, R, Aghajari, N. | | Deposit date: | 2009-02-19 | | Release date: | 2009-05-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural determinants of product specificity of sucrose isomerases
Febs Lett., 583, 2009
|
|
3GBD
 
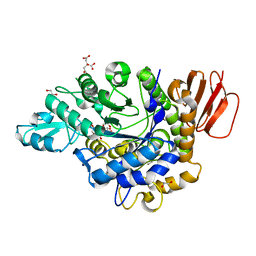 | | Crystal structure of the isomaltulose synthase SmuA from Protaminobacter rubrum | | Descriptor: | 1,2-ETHANEDIOL, CITRATE ANION, Sucrose isomerase SmuA from Protaminobacter rubrum | | Authors: | Ravaud, S, Robert, X, Haser, R, Aghajari, N. | | Deposit date: | 2009-02-19 | | Release date: | 2009-05-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural determinants of product specificity of sucrose isomerases
Febs Lett., 583, 2009
|
|
6RNH
 
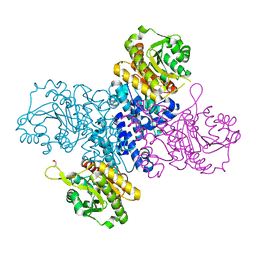 | | Structure of C-terminal truncated Plasmodium falciparum IMP-nucleotidase | | Descriptor: | GLYCEROL, IMP-specific 5'-nucleotidase, putative | | Authors: | Carrique, L, Ballut, L, Violot, S, Aghajari, N. | | Deposit date: | 2019-05-08 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structure and catalytic regulation of Plasmodium falciparum IMP specific nucleotidase.
Nat Commun, 11, 2020
|
|
6RN1
 
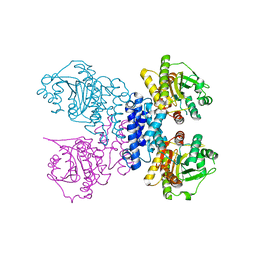 | | Structure of N-terminal truncated Plasmodium falciparum IMP-nucleotidase | | Descriptor: | IMP-specific 5'-nucleotidase, putative | | Authors: | Carrique, L, Ballut, L, Violot, S, Aghajari, N. | | Deposit date: | 2019-05-07 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure and catalytic regulation of Plasmodium falciparum IMP specific nucleotidase.
Nat Commun, 11, 2020
|
|
6RMO
 
 | | Structure of Plasmodium falciparum IMP-nucleotidase | | Descriptor: | IMP-specific 5'-nucleotidase, putative | | Authors: | Carrique, L, Ballut, L, Violot, S, Aghajari, N. | | Deposit date: | 2019-05-07 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and catalytic regulation of Plasmodium falciparum IMP specific nucleotidase.
Nat Commun, 11, 2020
|
|
6RME
 
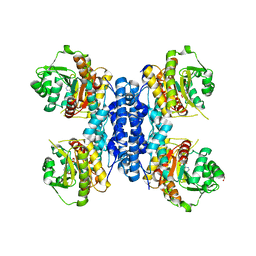 | | Structure of IMP bound Plasmodium falciparum IMP-nucleotidase mutant D172N | | Descriptor: | GLYCEROL, IMP-specific 5'-nucleotidase, putative, ... | | Authors: | Carrique, L, Ballut, L, Violot, S, Aghajari, N. | | Deposit date: | 2019-05-06 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure and catalytic regulation of Plasmodium falciparum IMP specific nucleotidase.
Nat Commun, 11, 2020
|
|
6RMD
 
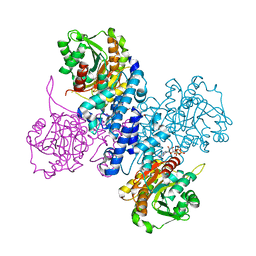 | | Structure of ATP bound Plasmodium falciparum IMP-nucleotidase | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, IMP-specific 5'-nucleotidase, ... | | Authors: | Carrique, L, Ballut, L, Violot, S, Aghajari, N. | | Deposit date: | 2019-05-06 | | Release date: | 2020-07-15 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure and catalytic regulation of Plasmodium falciparum IMP specific nucleotidase.
Nat Commun, 11, 2020
|
|
6RMW
 
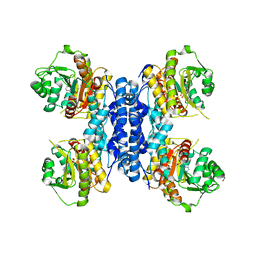 | | Structure of N-terminal truncated IMP bound Plasmodium falciparum IMP-nucleotidase | | Descriptor: | GLYCEROL, IMP-specific 5'-nucleotidase, putative, ... | | Authors: | Carrique, L, Ballut, L, Violot, S, Aghajari, N. | | Deposit date: | 2019-05-07 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure and catalytic regulation of Plasmodium falciparum IMP specific nucleotidase.
Nat Commun, 11, 2020
|
|
8R3S
 
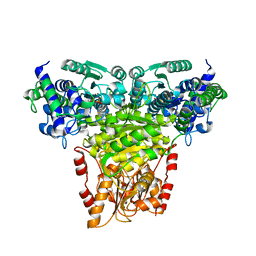 | | Transketolase from Staphylococcus aureus in complex with thiamin pyrophosphate | | Descriptor: | MAGNESIUM ION, THIAMINE DIPHOSPHATE, Transketolase | | Authors: | Ballut, L, Georges, N, Aghajari, N, Hecquet, L, Charmantray, F, Doumeche, B. | | Deposit date: | 2023-11-10 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Are Transketolases relevant Targets fighting Human Pathogens? A comparative Biochemical, Bioinformatic and Structural Study
To Be Published
|
|
1G87
 
 | | THE CRYSTAL STRUCTURE OF ENDOGLUCANASE 9G FROM CLOSTRIDIUM CELLULOLYTICUM | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, ENDOCELLULASE 9G, ... | | Authors: | Mandelman, D, Belaich, A, Belaich, J.P, Aghajari, N, Driguez, H, Haser, R. | | Deposit date: | 2000-11-16 | | Release date: | 2003-07-15 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-Ray Crystal Structure of the Multidomain Endoglucanase Cel9G from Clostridium
cellulolyticum Complexed with Natural and Synthetic Cello-Oligosaccharides
J.BACTERIOL., 185, 2003
|
|
1GA2
 
 | | THE CRYSTAL STRUCTURE OF ENDOGLUCANASE 9G FROM CLOSTRIDIUM CELLULOLYTICUM COMPLEXED WITH CELLOBIOSE | | Descriptor: | ACETIC ACID, CALCIUM ION, ENDOGLUCANASE 9G, ... | | Authors: | Mandelman, D, Belaich, A, Belaich, J.P, Aghajari, N, Driguez, H, Haser, R. | | Deposit date: | 2000-11-29 | | Release date: | 2003-07-22 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-Ray Crystal Structure of the Multidomain Endoglucanase Cel9G from Clostridium
cellulolyticum Complexed with Natural and Synthetic Cello-Oligosaccharides
J.BACTERIOL., 185, 2003
|
|
4D5U
 
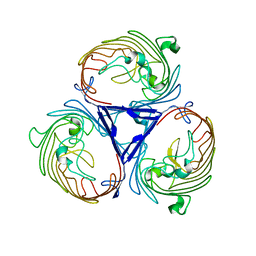 | | Structure of OmpF in I2 | | Descriptor: | OUTER MEMBRANE PROTEIN F | | Authors: | Chaptal, V, Kilburg, A, Flot, D, Wiseman, B, Aghajari, N, Jault, J.M, Falson, P. | | Deposit date: | 2014-11-07 | | Release date: | 2015-12-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Two Different Centered Monoclinic Crystals of the E. Coli Outer-Membrane Protein Ompf Originate from the Same Building Block.
Biochim.Biophys.Acta, 1858, 2016
|
|
7O7T
 
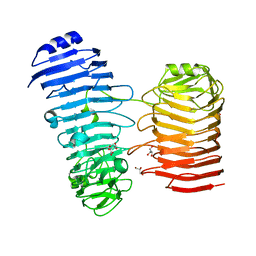 | | Structure of the PL6 family alginate lyase Patl3640 from Pseudoalteromonas atlantica T6c in complex with 4-deoxy-L-erythro-5-hexoseulose uronic acid | | Descriptor: | 1,2-ETHANEDIOL, 4-deoxy-L-erythro-hex-5-ulosuronic acid, GLYCEROL, ... | | Authors: | Ballut, L, Violot, S, Carrique, L, Aghajari, N. | | Deposit date: | 2021-04-13 | | Release date: | 2021-07-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Exploring molecular determinants of polysaccharide lyase family 6-1 enzyme activity.
Glycobiology, 31, 2021
|
|
7O84
 
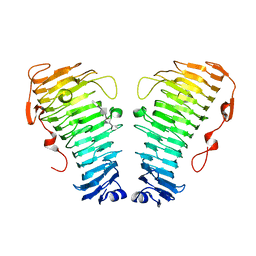 | | Structure of the PL6 family alginate lyase Pedsa0632 from Pseudopedobacter saltans in complex with substrate | | Descriptor: | 4-deoxy-alpha-L-erythro-hex-4-enopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid, 4-deoxy-alpha-L-erythro-hex-4-enopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid, Alginate lyase | | Authors: | Ballut, L, Violot, S, Carrique, L, Aghajari, N. | | Deposit date: | 2021-04-14 | | Release date: | 2021-07-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.177 Å) | | Cite: | Exploring molecular determinants of polysaccharide lyase family 6-1 enzyme activity.
Glycobiology, 31, 2021
|
|
7O77
 
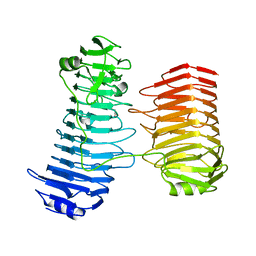 | | Structure of the PL6 family alginate lyase Patl3640 from Pseudoalteromonas atlantica T6c | | Descriptor: | GLYCEROL, Poly(Beta-D-mannuronate) lyase | | Authors: | Ballut, L, Violot, S, Carrique, L, Aghajari, N. | | Deposit date: | 2021-04-13 | | Release date: | 2021-07-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.321 Å) | | Cite: | Exploring molecular determinants of polysaccharide lyase family 6-1 enzyme activity.
Glycobiology, 31, 2021
|
|
7O78
 
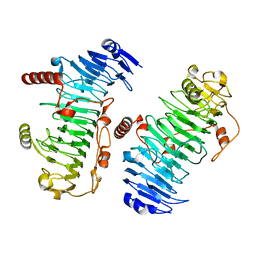 | | Structure of the PL6 family chondroitinase B from Pseudopedobacter saltans, Pedsa3807 | | Descriptor: | Polysaccharide lyase from Pseudopedobacter saltans, Pedsa3807 | | Authors: | Ballut, L, Violot, S, Carrique, L, Aghajari, N. | | Deposit date: | 2021-04-13 | | Release date: | 2021-07-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Exploring molecular determinants of polysaccharide lyase family 6-1 enzyme activity.
Glycobiology, 31, 2021
|
|
7O7A
 
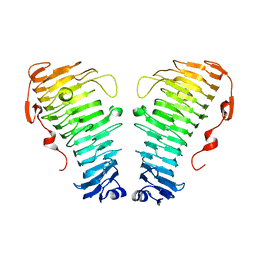 | |
7O79
 
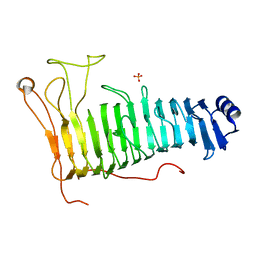 | | Structure of the PL6 family polysaccharide lyase Pedsa3628 from Pseudopedobacter saltans | | Descriptor: | PHOSPHATE ION, Poly(Beta-D-mannuronate) lyase | | Authors: | Ballut, L, Violot, S, Carrique, L, Aghajari, N. | | Deposit date: | 2021-04-13 | | Release date: | 2021-07-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Exploring molecular determinants of polysaccharide lyase family 6-1 enzyme activity.
Glycobiology, 31, 2021
|
|
1O0Q
 
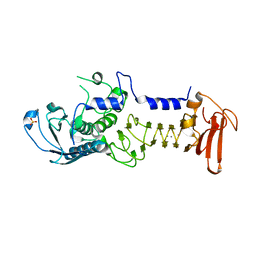 | | Crystal structure of a cold adapted alkaline protease from Pseudomonas TAC II 18, co-crystallized with 1 mM EDTA | | Descriptor: | CALCIUM ION, SULFATE ION, serralysin | | Authors: | Ravaud, S, Gouet, P, Haser, R, Aghajari, N. | | Deposit date: | 2003-02-24 | | Release date: | 2003-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Probing the role of divalent metal ions in a bacterial psychrophilic metalloprotease: binding studies of an enzyme in the crystalline state by x-ray crystallography.
J.Bacteriol., 185, 2003
|
|
1O0T
 
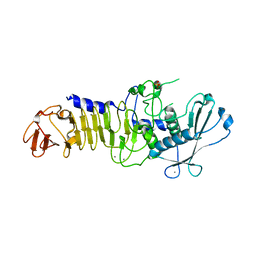 | | CRYSTAL STRUCTURE OF A COLD ADAPTED ALKALINE PROTEASE FROM PSEUDOMONAS TAC II 18, CO-CRYSTALLIZED WITH 5 mM EDTA (5 DAYS) | | Descriptor: | CALCIUM ION, SULFATE ION, serralysin | | Authors: | Ravaud, S, Gouet, P, Haser, R, Aghajari, N. | | Deposit date: | 2003-02-24 | | Release date: | 2003-07-15 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Probing the role of divalent metal ions in a bacterial psychrophilic metalloprotease: binding studies of an enzyme in the crystalline state by x-ray crystallography.
J.Bacteriol., 185, 2003
|
|
1OM8
 
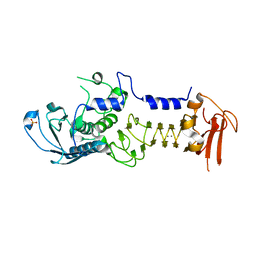 | | CRYSTAL STRUCTURE OF A COLD ADAPTED ALKALINE PROTEASE FROM PSEUDOMONAS TAC II 18, CO-CRYSTALLYZED WITH 10 mM EDTA | | Descriptor: | CALCIUM ION, SERRALYSIN, SULFATE ION | | Authors: | Ravaud, S, Gouet, P, Haser, R, Aghajari, N. | | Deposit date: | 2003-02-25 | | Release date: | 2003-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Probing the role of divalent metal ions in a bacterial psychrophilic metalloprotease: binding studies of an enzyme in the crystalline state by x-ray crystallography.
J.Bacteriol., 185, 2003
|
|
