2H5D
 
 | | 0.9A resolution crystal structure of alpha-lytic protease complexed with a transition state analogue, MeOSuc-Ala-Ala-Pro-Val boronic acid | | Descriptor: | ALPHA-LYTIC PROTEASE, GLYCEROL, MEOSUC-ALA-ALA-PRO-ALA BORONIC ACID INHIBITOR, ... | | Authors: | Fuhrmann, C.N, Agard, D.A. | | Deposit date: | 2006-05-25 | | Release date: | 2006-09-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Subangstrom crystallography reveals that short ionic hydrogen bonds, and not a His-Asp low-barrier hydrogen bond, stabilize the transition state in serine protease catalysis
J.Am.Chem.Soc., 128, 2006
|
|
1P01
 
 | |
1P10
 
 | |
1P09
 
 | |
1Z5V
 
 | | Crystal structure of human gamma-tubulin bound to GTPgammaS | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, MAGNESIUM ION, Tubulin gamma-1 chain | | Authors: | Aldaz, H.A, Rice, L.M, Stearns, T, Agard, D.A. | | Deposit date: | 2005-03-20 | | Release date: | 2005-05-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Insights into microtubule nucleation from the crystal structure of human gamma-tubulin.
Nature, 435, 2005
|
|
1LE4
 
 | |
1Z5W
 
 | | Crystal Structure of gamma-tubulin bound to GTP | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Tubulin gamma-1 chain | | Authors: | Aldaz, H.A, Rice, L.M, Stearns, T, Agard, D.A. | | Deposit date: | 2005-03-20 | | Release date: | 2005-05-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Insights into microtubule nucleation from the crystal structure of human gamma-tubulin.
Nature, 435, 2005
|
|
1LE2
 
 | |
5FM1
 
 | | Structure of gamma-tubulin small complex based on a cryo-EM map, chemical cross-links, and a remotely related structure | | Descriptor: | SPINDLE POLE BODY COMPONENT 110, SPINDLE POLE BODY COMPONENT SPC97, SPINDLE POLE BODY COMPONENT SPC98, ... | | Authors: | Greenberg, C.H, Kollman, J, Zelter, A, Johnson, R, MacCoss, M.J, Davis, T.N, Agard, D.A, Sali, A. | | Deposit date: | 2015-10-30 | | Release date: | 2016-02-03 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Structure of Gamma-Tubulin Small Complex Based on a Cryo-Em Map, Chemical Cross-Links, and a Remotely Related Structure.
J.Struct.Biol., 194, 2016
|
|
5FLZ
 
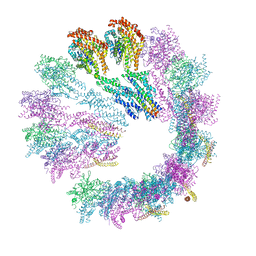 | | Cryo-EM structure of gamma-TuSC oligomers in a closed conformation | | Descriptor: | SPINDLE POLE BODY COMPONENT 110, SPINDLE POLE BODY COMPONENT SPC97, SPINDLE POLE BODY COMPONENT SPC98, ... | | Authors: | Greenberg, C.H, Kollman, J, Zelter, A, Johnson, R, MacCoss, M.J, Davis, T.N, Agard, D.A, Sali, A. | | Deposit date: | 2015-10-29 | | Release date: | 2016-01-13 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Structure of Gamma-Tubulin Small Complex Based on a Cryo-Em Map, Chemical Cross-Links, and a Remotely Related Structure.
J.Struct.Biol., 194, 2016
|
|
4PRO
 
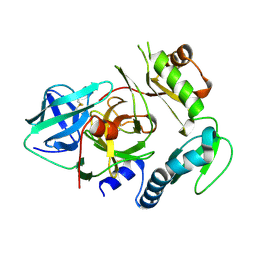 | |
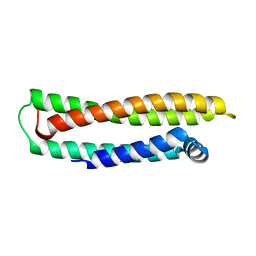
1LPE
 
 | |
1L2I
 
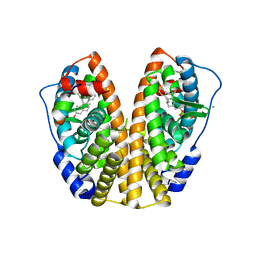 | | Human Estrogen Receptor alpha Ligand-binding Domain in Complex with (R,R)-5,11-cis-diethyl-5,6,11,12-tetrahydrochrysene-2,8-diol and a Glucocorticoid Receptor Interacting Protein 1 NR box II Peptide | | Descriptor: | (R,R)-5,11-CIS-DIETHYL-5,6,11,12-TETRAHYDROCHRYSENE-2,8-DIOL, CHLORIDE ION, ESTROGEN RECEPTOR, ... | | Authors: | Shiau, A.K, Barstad, D, Radek, J.T, Meyers, M.J, Nettles, K.W, Katzenellenbogen, B.S, Katzenellenbogen, J.A, Agard, D.A, Greene, G.L. | | Deposit date: | 2002-02-21 | | Release date: | 2002-05-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural characterization of a subtype-selective ligand reveals a novel mode of estrogen receptor antagonism.
Nat.Struct.Biol., 9, 2002
|
|
1SSX
 
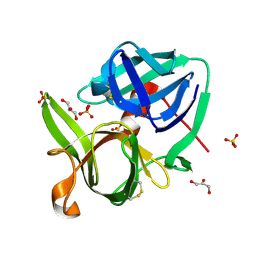 | |
1SF8
 
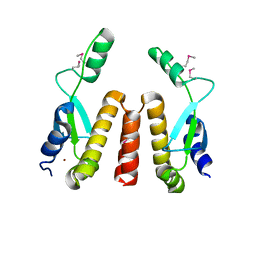 | | Crystal structure of the carboxy-terminal domain of htpG, the E. coli Hsp90 | | Descriptor: | CHLORIDE ION, Chaperone protein htpG, NICKEL (II) ION | | Authors: | Harris, S.F, Shiau, A.K, Agard, D.A. | | Deposit date: | 2004-02-19 | | Release date: | 2004-06-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The crystal structure of the carboxy-terminal dimerization domain of htpG, the Escherichia coli Hsp90, reveals a potential substrate binding site.
Structure, 12, 2004
|
|
7LPR
 
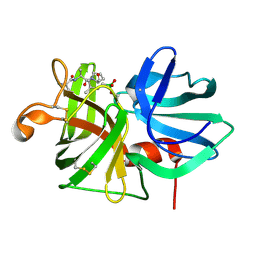 | |
3ERD
 
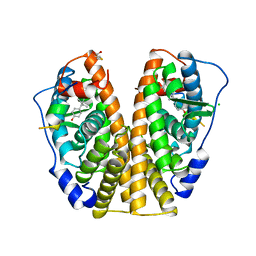 | | HUMAN ESTROGEN RECEPTOR ALPHA LIGAND-BINDING DOMAIN IN COMPLEX WITH DIETHYLSTILBESTROL AND A GLUCOCORTICOID RECEPTOR INTERACTING PROTEIN 1 NR BOX II PEPTIDE | | Descriptor: | ACETIC ACID, CHLORIDE ION, DIETHYLSTILBESTROL, ... | | Authors: | Shiau, A.K, Barstad, D, Loria, P.M, Cheng, L, Kushner, P.J, Agard, D.A, Greene, G.L. | | Deposit date: | 1999-03-31 | | Release date: | 1999-04-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | The structural basis of estrogen receptor/coactivator recognition and the antagonism of this interaction by tamoxifen.
Cell(Cambridge,Mass.), 95, 1998
|
|
9GDY
 
 | | SARS-CoV-2 Spike protein Beta Variant at 37C structural flexibility / heterogeneity analyses | | Descriptor: | Spike glycoprotein,Fibritin | | Authors: | Herreros, D, Mata, C.P, Noddings, C, Irene, D, Agard, D.A, Tsai, M.-D, Sorzano, C.O.S, Carazo, J.M. | | Deposit date: | 2024-08-06 | | Release date: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Real-space heterogeneous reconstruction, refinement, and disentanglement of CryoEM conformational states with HetSIREN.
Biorxiv, 2024
|
|
9GDX
 
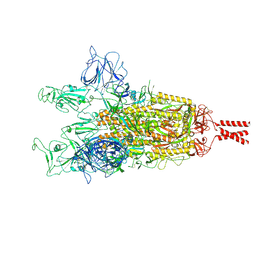 | | SARS-CoV-2 Spike protein Beta Variant at 4C structural flexibility / heterogeneity analyses | | Descriptor: | Spike glycoprotein,Fibritin | | Authors: | Herreros, D, Mata, C.P, Noddings, C, Irene, D, Agard, D.A, Tsai, M.-D, Sorzano, C.O.S, Carazo, J.M. | | Deposit date: | 2024-08-06 | | Release date: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Real-space heterogeneous reconstruction, refinement, and disentanglement of CryoEM conformational states with HetSIREN.
Biorxiv, 2024
|
|
3J4S
 
 | | Helical Model of TubZ-Bt four-stranded filament | | Descriptor: | FtsZ/tubulin-related protein, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Montabana, E.A, Agard, D.A. | | Deposit date: | 2013-10-03 | | Release date: | 2014-02-19 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Bacterial tubulin TubZ-Bt transitions between a two-stranded intermediate and a four-stranded filament upon GTP hydrolysis.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3ERT
 
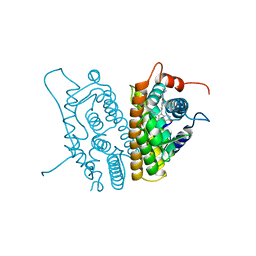 | | HUMAN ESTROGEN RECEPTOR ALPHA LIGAND-BINDING DOMAIN IN COMPLEX WITH 4-HYDROXYTAMOXIFEN | | Descriptor: | 4-HYDROXYTAMOXIFEN, PROTEIN (ESTROGEN RECEPTOR ALPHA) | | Authors: | Shiau, A.K, Barstad, D, Loria, P.M, Cheng, L, Kushner, P.J, Agard, D.A, Greene, G.L. | | Deposit date: | 1999-03-30 | | Release date: | 1999-04-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structural basis of estrogen receptor/coactivator recognition and the antagonism of this interaction by tamoxifen.
Cell(Cambridge,Mass.), 95, 1998
|
|
3J9I
 
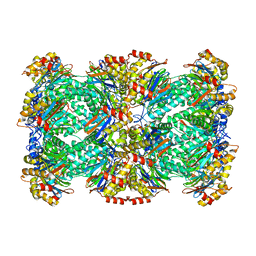 | | Thermoplasma acidophilum 20S proteasome | | Descriptor: | Proteasome subunit alpha, Proteasome subunit beta | | Authors: | Li, X, Mooney, P, Zheng, S, Booth, C, Braunfeld, M.B, Gubbens, S, Agard, D.A, Cheng, Y. | | Deposit date: | 2015-02-02 | | Release date: | 2015-02-18 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM.
Nat.Methods, 10, 2013
|
|
4IPE
 
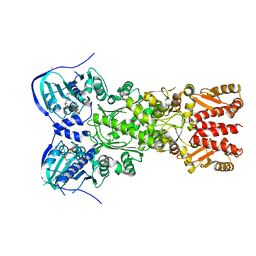 | | Crystal structure of mitochondrial Hsp90 (TRAP1) with AMPPNP | | Descriptor: | COBALT (II) ION, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Partridge, J.R, Lavery, L.A, Agard, D.A. | | Deposit date: | 2013-01-09 | | Release date: | 2014-01-22 | | Last modified: | 2014-08-27 | | Method: | X-RAY DIFFRACTION (2.289 Å) | | Cite: | Structural asymmetry in the closed state of mitochondrial Hsp90 (TRAP1) supports a two-step ATP hydrolysis mechanism.
Mol.Cell, 53, 2014
|
|
4J0B
 
 | | Structure of mitochondrial Hsp90 (TRAP1) with ADP-BeF3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, COBALT (II) ION, ... | | Authors: | Partridge, J.R, Lavery, L.A, Agard, D.A. | | Deposit date: | 2013-01-30 | | Release date: | 2014-01-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.352 Å) | | Cite: | Structural asymmetry in the closed state of mitochondrial Hsp90 (TRAP1) supports a two-step ATP hydrolysis mechanism.
Mol.Cell, 53, 2014
|
|
4IVG
 
 | | Crystal structure of mitochondrial Hsp90 (TRAP1) NTD-Middle domain dimer with AMPPNP | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Partridge, J.R, Lavery, L.A, Agard, D.A. | | Deposit date: | 2013-01-22 | | Release date: | 2014-01-22 | | Last modified: | 2014-08-27 | | Method: | X-RAY DIFFRACTION (1.749 Å) | | Cite: | Structural asymmetry in the closed state of mitochondrial Hsp90 (TRAP1) supports a two-step ATP hydrolysis mechanism.
Mol.Cell, 53, 2014
|
|
