3M7T
 
 | |
3M7U
 
 | |
3J4T
 
 | | Helical model of TubZ-Bt two-stranded filament | | Descriptor: | FtsZ/tubulin-related protein | | Authors: | Agard, D.A, Montabana, E.A. | | Deposit date: | 2013-10-04 | | Release date: | 2014-02-19 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (10.8 Å) | | Cite: | Bacterial tubulin TubZ-Bt transitions between a two-stranded intermediate and a four-stranded filament upon GTP hydrolysis.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3R4V
 
 | | Structure of the phage tubulin PhuZ-GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Putative uncharacterized protein | | Authors: | Agard, D.A, Pogliano, J, Kraemer, J.A, Erb, M.L, Waddling, C.A, Montabana, E.A, Wang, H, Nguyen, K, Pham, S. | | Deposit date: | 2011-03-17 | | Release date: | 2012-07-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | A phage tubulin assembles dynamic filaments by an atypical mechanism to center viral DNA within the host cell.
Cell(Cambridge,Mass.), 149, 2012
|
|
3RB8
 
 | | Structure of the phage tubulin PhuZ(SeMet)-GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Putative uncharacterized protein | | Authors: | Agard, D.A, Pogliano, J, Kraemer, J.A, Erb, M.L, Waddling, C.A, Montabana, E.A, Wang, H, Nguyen, K, Pham, S. | | Deposit date: | 2011-03-28 | | Release date: | 2012-07-04 | | Last modified: | 2013-01-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A phage tubulin assembles dynamic filaments by an atypical mechanism to center viral DNA within the host cell.
Cell(Cambridge,Mass.), 149, 2012
|
|
8LPR
 
 | |
5LPR
 
 | |
9LPR
 
 | |
6D14
 
 | |
3URE
 
 | |
3URC
 
 | | T181G mutant of alpha-Lytic Protease | | Descriptor: | Alpha-lytic protease, GLYCEROL, SULFATE ION | | Authors: | Kelch, B.A, Agard, D.A. | | Deposit date: | 2011-11-22 | | Release date: | 2012-05-23 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Functional modulation of a protein folding landscape via side-chain distortion.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
6LPR
 
 | |
5WP9
 
 | | Structural Basis of Mitochondrial Receptor Binding and Constriction by Dynamin-Related Protein 1 | | Descriptor: | Dynamin-1-like protein, MAGNESIUM ION, Mitochondrial dynamics protein MID49, ... | | Authors: | Kalia, R, Wang, R.Y.R, Yusuf, A, Thomas, P.V, Agard, D.A, Shaw, J.M, Frost, A. | | Deposit date: | 2017-08-03 | | Release date: | 2018-06-20 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.22 Å) | | Cite: | Structural basis of mitochondrial receptor binding and constriction by DRP1.
Nature, 558, 2018
|
|
3URD
 
 | | T181A mutant of alpha-Lytic Protease | | Descriptor: | Alpha-lytic protease, GLYCEROL, SULFATE ION | | Authors: | Kelch, B.A, Agard, D.A. | | Deposit date: | 2011-11-22 | | Release date: | 2012-05-23 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Functional modulation of a protein folding landscape via side-chain distortion.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1BOQ
 
 | | PRO REGION C-TERMINUS: PROTEASE ACTIVE SITE INTERACTIONS ARE CRITICAL IN CATALYZING THE FOLDING OF ALPHA-LYTIC PROTEASE | | Descriptor: | PROTEIN (ALPHA-LYTIC PROTEASE), SULFATE ION | | Authors: | Peters, R.J, Shiau, A.K, Sohl, J.L, Anderson, D.E, Tang, G, Silen, J.L, Agard, D.A. | | Deposit date: | 1998-08-05 | | Release date: | 1998-08-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Pro region C-terminus:protease active site interactions are critical in catalyzing the folding of alpha-lytic protease.
Biochemistry, 37, 1998
|
|
2ULL
 
 | |
3CB2
 
 | |
3LPR
 
 | |
6XG6
 
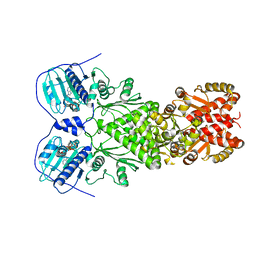 | | Full-length human mitochondrial Hsp90 (TRAP1) with ADP-BeF3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Heat shock protein 75 kDa, ... | | Authors: | Liu, Y.X, Wang, F, Agard, D.A. | | Deposit date: | 2020-06-17 | | Release date: | 2020-09-30 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | General and robust covalently linked graphene oxide affinity grids for high-resolution cryo-EM.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6XLC
 
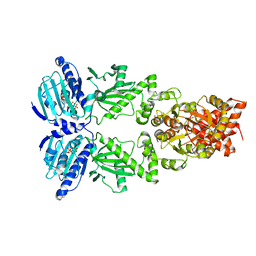 | | Full-length Hsc82 bound to AMPPNP | | Descriptor: | ATP-dependent molecular chaperone HSC82, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Liu, Y.X, Sun, M, Myasnikov, A.G, Elnatan, D, Agard, D.A. | | Deposit date: | 2020-06-28 | | Release date: | 2021-06-30 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | Full-length Hsc82 bound to AMPPNP
To Be Published
|
|
6XLB
 
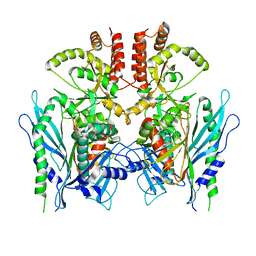 | | Apo full-length Hsc82 in complex with Aha1 | | Descriptor: | ATP-dependent molecular chaperone HSC82, Hsp90 co-chaperone AHA1 | | Authors: | Liu, Y.X, Sun, M, Myasnikov, A.G, Elnatan, D, Agard, D.A. | | Deposit date: | 2020-06-28 | | Release date: | 2021-06-30 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Apo full-length Hsc82 in complex with Aha1
To Be Published
|
|
6XLE
 
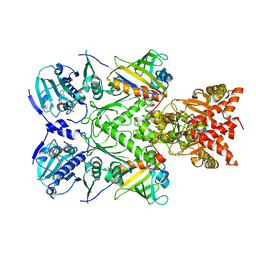 | | Full-length Hsc82 in complex with two Aha1 CTD in the presence of AMP-PNP | | Descriptor: | ATP-dependent molecular chaperone HSC82, Hsp90 co-chaperone AHA1, MAGNESIUM ION, ... | | Authors: | Liu, Y.X, Sun, M, Myasnikov, A.G, Elnatan, D, Agard, D.A. | | Deposit date: | 2020-06-28 | | Release date: | 2021-06-30 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | Cryo-EM structures reveal a multistep mechanism of Hsp90 activation by co-chaperone Aha1
To Be Published
|
|
6XLF
 
 | | Full-length Hsc82 in complex with Aha1 in the presence of AMP-PNP | | Descriptor: | ATP-dependent molecular chaperone HSC82, Hsp90 co-chaperone AHA1, MAGNESIUM ION, ... | | Authors: | Liu, Y.X, Sun, M, Myasnikov, A.G, Elnatan, D, Agard, D.A. | | Deposit date: | 2020-06-28 | | Release date: | 2021-06-30 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Cryo-EM structures reveal a multistep mechanism of Hsp90 activation by co-chaperone Aha1
To Be Published
|
|
6XLG
 
 | | Full-length Hsc82 in complex with two Aha1 CTD in the presence of ATPgammaS | | Descriptor: | ATP-dependent molecular chaperone HSC82, Hsp90 co-chaperone AHA1, MAGNESIUM ION, ... | | Authors: | Liu, Y.X, Sun, M, Myasnikov, A.G, Elnatan, D, Agard, D.A. | | Deposit date: | 2020-06-28 | | Release date: | 2021-06-30 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | Cryo-EM structures reveal a multistep mechanism of Hsp90 activation by co-chaperone Aha1
To Be Published
|
|
6XLH
 
 | | Asymmetric hydrolysis state of Hsc82 in complex with Aha1 bound with ADP and ATPgammaS | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent molecular chaperone HSC82, Hsp90 co-chaperone AHA1, ... | | Authors: | Liu, Y.X, Sun, M, Myasnikov, A.G, Elnatan, D, Agard, D.A. | | Deposit date: | 2020-06-28 | | Release date: | 2021-06-30 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Cryo-EM structures reveal a multistep mechanism of Hsp90 activation by co-chaperone Aha1
To Be Published
|
|
