2Q8V
 
 | | NblA protein from T. vulcanus crystallized with urea | | Descriptor: | NblA protein, UREA | | Authors: | Adir, N, Dines, M. | | Deposit date: | 2007-06-12 | | Release date: | 2008-07-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural, Functional, and Mutational Analysis of the NblA Protein Provides Insight into Possible Modes of Interaction with the Phycobilisome
J.Biol.Chem., 283, 2008
|
|
4D87
 
 | |
1KTP
 
 | | Crystal structure of c-phycocyanin of synechococcus vulcanus at 1.6 angstroms | | Descriptor: | BILIVERDINE IX ALPHA, C-PHYCOCYANIN ALPHA SUBUNIT, C-PHYCOCYANIN BETA SUBUNIT, ... | | Authors: | Adir, N, Dobrovetsky, E, Lerner, N. | | Deposit date: | 2002-01-17 | | Release date: | 2002-03-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Refined structure of c-phycocyanin from the cyanobacterium Synechococcus vulcanus at 1.6 A: insights into the role of solvent molecules in thermal stability and co-factor structure
Biochim.Biophys.Acta, 1556, 2002
|
|
6EWN
 
 | | HspA from Thermosynechococcus vulcanus in the presence of 2M urea with initial stages of denaturation | | Descriptor: | HspA, UREA | | Authors: | Adir, N, Ghosh, S, Salama, F, Dines, M. | | Deposit date: | 2017-11-06 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Biophysical and structural characterization of the small heat shock protein HspA from Thermosynechococcus vulcanus in 2 M urea.
Biochim Biophys Acta Proteins Proteom, 1867, 2019
|
|
1I7Y
 
 | |
3DBJ
 
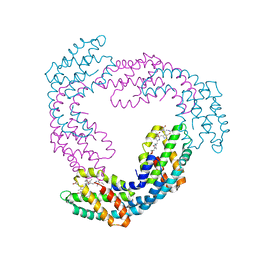 | | Allophycocyanin from Thermosynechococcus vulcanus | | Descriptor: | Allophycocyanin, PHYCOCYANOBILIN | | Authors: | Adir, N, Klartag, M, McGregor, A, David, L. | | Deposit date: | 2008-06-01 | | Release date: | 2008-11-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Allophycocyanin Trimer Stability and Functionality Are Primarily Due to Polar Enhanced Hydrophobicity of the Phycocyanobilin Binding Pocket
J.Mol.Biol., 384, 2008
|
|
1ON7
 
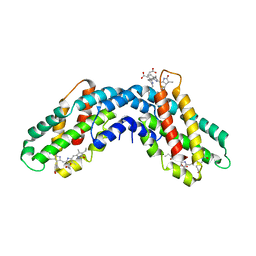 | |
5OOK
 
 | | Structure of A. marina Phycocyanin contains overlapping isoforms | | Descriptor: | DI(HYDROXYETHYL)ETHER, PHYCOCYANOBILIN, Phycocyanin, ... | | Authors: | Bar-Zvi, S, Lahav, A, Blankenship, E.R, Adir, N. | | Deposit date: | 2017-08-08 | | Release date: | 2018-06-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural heterogeneity leads to functional homogeneity in A. marina phycocyanin.
Biochim. Biophys. Acta, 1859, 2018
|
|
8QX5
 
 | | Helical Carotenoid Protein 4 (HCP4) from Anabaena with bound Canthaxanthin | | Descriptor: | Orange carotenoid-binding domain-containing protein, beta,beta-carotene-4,4'-dione | | Authors: | Sklyar, J, Wilson, A, Kirilovsky, D, Adir, N. | | Deposit date: | 2023-10-22 | | Release date: | 2024-04-03 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Insights into energy quenching mechanisms and carotenoid uptake by orange carotenoid protein homologs: HCP4 and CTDH.
Int.J.Biol.Macromol., 265, 2024
|
|
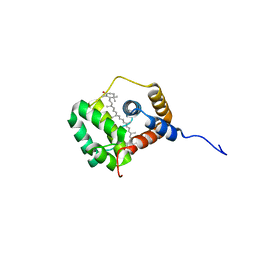
8QX7
 
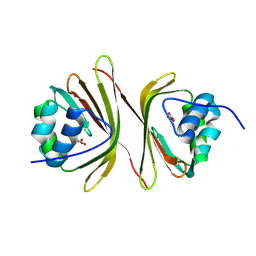 | | Apo-C-Terminal Domain Homolog of the Orange Carotenoid Protein from Anabaena at a resolution of 1.95 Angstroms | | Descriptor: | All4940 protein, MALONIC ACID | | Authors: | Sklyar, J, Wilson, A, Kirilovsky, D, Adir, N. | | Deposit date: | 2023-10-22 | | Release date: | 2024-04-03 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Insights into energy quenching mechanisms and carotenoid uptake by orange carotenoid protein homologs: HCP4 and CTDH.
Int.J.Biol.Macromol., 265, 2024
|
|
2QDO
 
 | | NblA protein from T. vulcanus | | Descriptor: | NblA protein | | Authors: | Dines, M, Adir, N. | | Deposit date: | 2007-06-21 | | Release date: | 2008-07-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural, Functional, and Mutational Analysis of the NblA Protein Provides Insight into Possible Modes of Interaction with the Phycobilisome
J.Biol.Chem., 283, 2008
|
|
6S5L
 
 | |
8OZ4
 
 | | Populus tremula stable protein 1 with an alternate crystal lattice | | Descriptor: | Stable protein 1 | | Authors: | Sklyar, J, Zeibaq, Y, Bachar, O, Yehezkeli, O, Adir, N. | | Deposit date: | 2023-05-08 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A Bioengineered Stable Protein 1-Hemin Complex with Enhanced Peroxidase-Like Catalytic Properties
Small Science, 2024
|
|
3UJP
 
 | | Structure of MntC protein at 2.7A | | Descriptor: | CACODYLATE ION, MANGANESE (II) ION, Mn transporter subunit, ... | | Authors: | Kanteev, M, Adir, N. | | Deposit date: | 2011-11-08 | | Release date: | 2012-11-14 | | Last modified: | 2013-06-19 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Arginine 116 stabilizes the entrance to the metal ion-binding site of the MntC protein.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
4ZC7
 
 | | Paromomycin bound to a leishmanial ribosomal A-site | | Descriptor: | PAROMOMYCIN, RNA duplex | | Authors: | Shalev, M, Rozenberg, H, Jaffe, C.L, Adir, N, Baasov, T. | | Deposit date: | 2015-04-15 | | Release date: | 2015-08-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.041 Å) | | Cite: | Structural basis for selective targeting of leishmanial ribosomes: aminoglycoside derivatives as promising therapeutics.
Nucleic Acids Res., 43, 2015
|
|
8OZS
 
 | | Populus tremula stable protein 1 with N-terminal binding peptide extension with hemin | | Descriptor: | Stable protein 1 | | Authors: | Sklyar, J, Zeibaq, Y, Bachar, O, Yehezkeli, O, Adir, N. | | Deposit date: | 2023-05-09 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A Bioengineered Stable Protein 1-Hemin Complex with Enhanced Peroxidase-Like Catalytic Properties
Small Science, 2023
|
|
8OZO
 
 | | Populus tremula stable protein 1 with N-terminal binding peptide extension | | Descriptor: | Stable protein 1 | | Authors: | Sklyar, J, Zeibaq, Y, Bachar, O, Yehezkeli, O, Adir, N. | | Deposit date: | 2023-05-09 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A Bioengineered Stable Protein 1-Hemin Complex with Enhanced Peroxidase-Like Catalytic Properties
Small Science, 2024
|
|
3CS5
 
 | | NblA protein from Synechococcus elongatus PCC 7942 | | Descriptor: | Phycobilisome degradation protein nblA | | Authors: | Dines, M, Sendersky, E, Schwarz, R, Adir, N. | | Deposit date: | 2008-04-09 | | Release date: | 2008-09-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural, Functional, and Mutational Analysis of the NblA Protein Provides Insight into Possible Modes of Interaction with the Phycobilisome
J.Biol.Chem., 283, 2008
|
|
6FEJ
 
 | | Anabaena Apo-C-Terminal Domain Homolog Protein | | Descriptor: | All4940 protein, UREA | | Authors: | Harris, D, Wilson, A, Muzzopappa, F, Kirilovsky, D, Adir, N. | | Deposit date: | 2018-01-02 | | Release date: | 2018-07-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural rearrangements in the C-terminal domain homolog of Orange Carotenoid Protein are crucial for carotenoid transfer.
Commun Biol, 1, 2018
|
|
1G7V
 
 | | CRYSTAL STRUCTURES OF KDO8P SYNTHASE IN ITS BINARY COMPLEXES WITH THE MECHANISM-BASED INHIBITOR | | Descriptor: | 2-DEHYDRO-3-DEOXYPHOSPHOOCTONATE ALDOLASE, {[(2,2-DIHYDROXY-ETHYL)-(2,3,4,5-TETRAHYDROXY-6-PHOSPHONOOXY-HEXYL)-AMINO]-METHYL}-PHOSPHONIC ACID | | Authors: | Asojo, O.A, Friedman, J.M, Belakhov, V, Shoham, Y, Adir, N, Baasov, T. | | Deposit date: | 2000-11-14 | | Release date: | 2001-05-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of KDOP synthase in its binary complexes with the substrate phosphoenolpyruvate and with a mechanism-based inhibitor.
Biochemistry, 40, 2001
|
|
1G7U
 
 | | CRYSTAL STRUCTURES OF KDO8P SYNTHASE IN ITS BINARY COMPLEX WITH SUBSTRATE PHOSPHOENOL PYRUVATE | | Descriptor: | 2-DEHYDRO-3-DEOXYPHOSPHOOCTONATE ALDOLASE, PHOSPHOENOLPYRUVATE | | Authors: | Asojo, O.A, Friedman, J.M, Belakhov, V, Shoham, Y, Adir, N, Baasov, T. | | Deposit date: | 2000-11-14 | | Release date: | 2001-05-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of KDOP synthase in its binary complexes with the substrate phosphoenolpyruvate and with a mechanism-based inhibitor.
Biochemistry, 40, 2001
|
|
3NTM
 
 | | Crystal Structure of Tyrosinase from Bacillus megaterium crystallized in the absence of zinc, partial occupancy of CuB | | Descriptor: | COPPER (II) ION, Tyrosinase | | Authors: | Sendovski, M, Kanteev, M, Adir, N, Fishman, A. | | Deposit date: | 2010-07-05 | | Release date: | 2010-11-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | First structures of an active bacterial tyrosinase reveal copper plasticity
J.Mol.Biol., 405, 2011
|
|
3NPY
 
 | | Crystal Structure of Tyrosinase from Bacillus megaterium soaked in CuSO4 | | Descriptor: | CHLORIDE ION, COPPER (II) ION, Tyrosinase, ... | | Authors: | Sendovski, M, Kanteev, M, Adir, N, Fishman, A. | | Deposit date: | 2010-06-29 | | Release date: | 2010-11-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.192 Å) | | Cite: | First structures of an active bacterial tyrosinase reveal copper plasticity.
J.Mol.Biol., 405, 2011
|
|
3NM8
 
 | | Crystal structure of Tyrosinase from Bacillus megaterium | | Descriptor: | CHLORIDE ION, COPPER (II) ION, Tyrosinase, ... | | Authors: | Sendovski, M, Kanteev, M, Adir, N, Fishman, A. | | Deposit date: | 2010-06-22 | | Release date: | 2010-11-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | First structures of an active bacterial tyrosinase reveal copper plasticity
J.Mol.Biol., 405, 2011
|
|
4K32
 
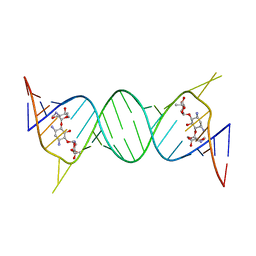 | | Crystal structure of geneticin bound to the leishmanial rRNA A-site | | Descriptor: | GENETICIN, RNA (5'-R(*UP*UP*GP*CP*GP*UP*CP*GP*UP*UP*CP*CP*GP*GP*AP*AP*AP*AP*GP*UP*CP*GP*C)-3') | | Authors: | Shalev, M, Kondo, J, Adir, N, Baasov, T. | | Deposit date: | 2013-04-10 | | Release date: | 2013-07-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Identification of the molecular attributes required for aminoglycoside activity against Leishmania.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
