6MWX
 
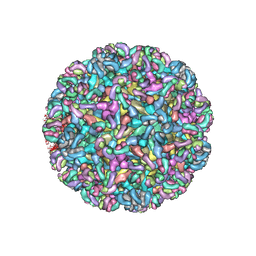 | | CryoEM structure of Chimeric Eastern Equine Encephalitis Virus with Fab of EEEV-69 Antibody | | Descriptor: | E1, E2, EEEV-69 antibody heavy chain, ... | | Authors: | Hasan, S.S, Sun, C, Kim, A.S, Watanabe, Y, Chen, C.L, Klose, T, Buda, G, Crispin, M, Diamond, M.S, Klimstra, W.B, Rossmann, M.G. | | Deposit date: | 2018-10-30 | | Release date: | 2018-12-19 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (8.2 Å) | | Cite: | Cryo-EM Structures of Eastern Equine Encephalitis Virus Reveal Mechanisms of Virus Disassembly and Antibody Neutralization.
Cell Rep, 25, 2018
|
|
1J1I
 
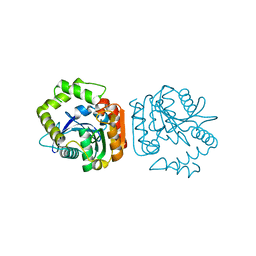 | | Crystal structure of a His-tagged Serine Hydrolase Involved in the Carbazole Degradation (CarC enzyme) | | Descriptor: | meta cleavage compound hydrolase | | Authors: | Habe, H, Morii, K, Fushinobu, S, Nam, J.W, Ayabe, Y, Yoshida, T, Wakagi, T, Yamane, H, Nojiri, H, Omori, T. | | Deposit date: | 2002-12-05 | | Release date: | 2003-06-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal structure of a histidine-tagged serine hydrolase involved in the carbazole degradation (CarC enzyme).
Biochem.Biophys.Res.Commun., 303, 2003
|
|
6MX7
 
 | | CryoEM structure of chimeric Eastern Equine Encephalitis Virus: Genome-Binding Capsid N-terminal Domain | | Descriptor: | Capsid | | Authors: | Hasan, S.S, Sun, C, Kim, A.S, Watanabe, Y, Chen, C.L, Klose, T, Buda, G, Crispin, M, Diamond, M.S, Klimstra, W.B, Rossmann, M.G. | | Deposit date: | 2018-10-30 | | Release date: | 2018-12-19 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Cryo-EM Structures of Eastern Equine Encephalitis Virus Reveal Mechanisms of Virus Disassembly and Antibody Neutralization.
Cell Rep, 25, 2018
|
|
6MX4
 
 | | CryoEM structure of chimeric Eastern Equine Encephalitis Virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Capsid, ... | | Authors: | Hasan, S.S, Sun, C, Kim, A.S, Watanabe, Y, Chen, C.L, Klose, T, Buda, G, Crispin, M, Diamond, M.S, Klimstra, W.B, Rossmann, M.G. | | Deposit date: | 2018-10-30 | | Release date: | 2018-12-19 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Cryo-EM Structures of Eastern Equine Encephalitis Virus Reveal Mechanisms of Virus Disassembly and Antibody Neutralization.
Cell Rep, 25, 2018
|
|
2ZX0
 
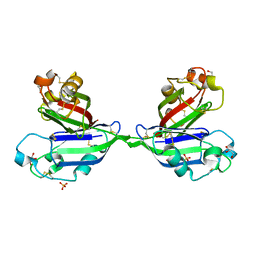 | | Rhamnose-binding lectin CSL3 | | Descriptor: | CSL3, GLYCEROL, PHOSPHATE ION | | Authors: | Shirai, T, Watababe, Y, Lee, M, Ogawa, T, Muramoto, K. | | Deposit date: | 2008-12-19 | | Release date: | 2009-06-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of rhamnose-binding lectin CSL3: unique pseudo-tetrameric architecture of a pattern recognition protein
J.Mol.Biol., 391, 2009
|
|
2ZX2
 
 | | Rhamnose-binding lectin CSL3 | | Descriptor: | CSL3, PHOSPHATE ION, alpha-L-rhamnopyranose | | Authors: | Shirai, T, Watababe, Y, Lee, M, Ogawa, T, Muramoto, K. | | Deposit date: | 2008-12-19 | | Release date: | 2009-06-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of rhamnose-binding lectin CSL3: unique pseudo-tetrameric architecture of a pattern recognition protein
J.Mol.Biol., 391, 2009
|
|
2ZX1
 
 | | Rhamnose-binding lectin CSL3 | | Descriptor: | CSL3, PHOSPHATE ION | | Authors: | Shirai, T, Watababe, Y, Lee, M, Ogawa, T, Muramoto, K. | | Deposit date: | 2008-12-19 | | Release date: | 2009-06-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of rhamnose-binding lectin CSL3: unique pseudo-tetrameric architecture of a pattern recognition protein
J.Mol.Biol., 391, 2009
|
|
2ZX4
 
 | | Rhamnose-binding lectin CSL3 | | Descriptor: | CSL3, PHOSPHATE ION, alpha-D-galactopyranose-(1-4)-beta-D-galactopyranose, ... | | Authors: | Shirai, T, Watababe, Y, Lee, M, Ogawa, T, Muramoto, K. | | Deposit date: | 2008-12-19 | | Release date: | 2009-06-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of rhamnose-binding lectin CSL3: unique pseudo-tetrameric architecture of a pattern recognition protein
J.Mol.Biol., 391, 2009
|
|
7F4Z
 
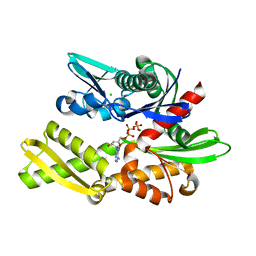 | | X-ray crystal structure of Y149A mutated Hsp72-NBD in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, Heat shock 70 kDa protein 1B, ... | | Authors: | Yokoyama, T, Fujii, S, Nabeshima, Y, Mizuguchi, M. | | Deposit date: | 2021-06-21 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Neutron crystallographic analysis of the nucleotide-binding domain of Hsp72 in complex with ADP.
Iucrj, 9, 2022
|
|
7F50
 
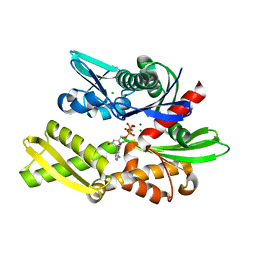 | | X-ray crystal structure of Y149A mutated Hsp72-NBD in complex with AMPPnP | | Descriptor: | CHLORIDE ION, Heat shock 70 kDa protein 1B, MAGNESIUM ION, ... | | Authors: | Yokoyama, T, Fujii, S, Nabeshima, Y, Mizuguchi, M. | | Deposit date: | 2021-06-21 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.703 Å) | | Cite: | Neutron crystallographic analysis of the nucleotide-binding domain of Hsp72 in complex with ADP.
Iucrj, 9, 2022
|
|
2ZX3
 
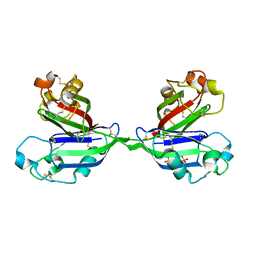 | | Rhamnose-binding lectin CSL3 | | Descriptor: | CSL3, PHOSPHATE ION, alpha-D-galactopyranose-(1-6)-beta-D-glucopyranose | | Authors: | Shirai, T, Watababe, Y, Lee, M, Ogawa, T, Muramoto, K. | | Deposit date: | 2008-12-19 | | Release date: | 2009-06-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of rhamnose-binding lectin CSL3: unique pseudo-tetrameric architecture of a pattern recognition protein
J.Mol.Biol., 391, 2009
|
|
4WR5
 
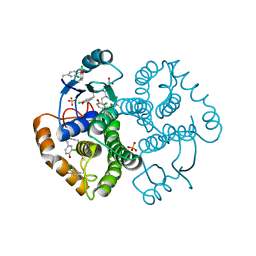 | | Crystal Structure of GST Mutated with Halogenated Tyrosine (7cGST-1) | | Descriptor: | GLUTATHIONE, Glutathione S-transferase class-mu 26 kDa isozyme, SULFATE ION | | Authors: | Akasaka, R, Kawazoe, M, Tomabechi, Y, Ohtake, K, Itagaki, T, Takemoto, C, Shirouzu, M, Yokoyama, S, Sakamoto, K. | | Deposit date: | 2014-10-23 | | Release date: | 2015-08-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Protein stabilization utilizing a redefined codon
Sci Rep, 5, 2015
|
|
5Y90
 
 | | MAP2K7 mutant -C218S | | Descriptor: | Dual specificity mitogen-activated protein kinase kinase 7, GLYCEROL | | Authors: | Kinoshita, T, Hashimoto, T, Sogabe, Y, Matsumoto, T, Sawa, M, Fukada, H. | | Deposit date: | 2017-08-22 | | Release date: | 2017-10-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | High-resolution structure discloses the potential for allosteric regulation of mitogen-activated protein kinase kinase 7
Biochem. Biophys. Res. Commun., 493, 2017
|
|
4WR4
 
 | | Crystal Structure of GST Mutated with Halogenated Tyrosine (7bGST-1) | | Descriptor: | GLUTATHIONE, Glutathione S-transferase class-mu 26 kDa isozyme, SULFATE ION | | Authors: | Akasaka, R, Kawazoe, M, Tomabechi, Y, Ohtake, K, Itagaki, T, Takemoto, C, Shirouzu, M, Yokoyama, S, Sakamoto, K. | | Deposit date: | 2014-10-23 | | Release date: | 2015-08-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Protein stabilization utilizing a redefined codon
Sci Rep, 5, 2015
|
|
7ARN
 
 | | Crystal Structure of the Fab Fragment of a Glycosylated Lymphoma Antibody | | Descriptor: | Antibody Fab Fragment Heavy Chain, Antibody Fab Fragment Light Chain, GLYCEROL, ... | | Authors: | Pryce, R, Allen, J.D, Watanabe, Y, Crispin, M, Bowden, T.A. | | Deposit date: | 2020-10-25 | | Release date: | 2021-11-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Crystal Structure of the Fab Fragment of a Glycosylated Lymphoma Antibody
To Be Published
|
|
8I75
 
 | |
8I76
 
 | |
8I74
 
 | |
6AJV
 
 | | Crystal structure of BRD4 in complex with isoliquiritigenin and DMSO (Cocktail No. 3) | | Descriptor: | 2',4,4'-TRIHYDROXYCHALCONE, Bromodomain-containing protein 4, DIMETHYL SULFOXIDE, ... | | Authors: | Yokoyama, T, Matsumoto, K, Nabeshima, Y, Mizuguchi, M. | | Deposit date: | 2018-08-28 | | Release date: | 2019-06-12 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural and thermodynamic characterization of the binding of isoliquiritigenin to the first bromodomain of BRD4.
Febs J., 286, 2019
|
|
6AJX
 
 | | Crystal structure of BRD4 in complex with isoliquiritigenin in the absence of DMSO | | Descriptor: | 2',4,4'-TRIHYDROXYCHALCONE, Bromodomain-containing protein 4, SODIUM ION | | Authors: | Yokoyama, T, Matsumoto, K, Nabeshima, Y, Mizuguchi, M. | | Deposit date: | 2018-08-28 | | Release date: | 2019-06-12 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.887 Å) | | Cite: | Structural and thermodynamic characterization of the binding of isoliquiritigenin to the first bromodomain of BRD4.
Febs J., 286, 2019
|
|
6AJY
 
 | | Crystal structure of BRD4 in complex with 2',4'-dihydroxy-2-methoxychalcone | | Descriptor: | 2',4'-dihydroxy-2-methoxychalcone, Bromodomain-containing protein 4, SODIUM ION | | Authors: | Yokoyama, T, Matsumoto, K, Nabeshima, Y, Mizuguchi, M. | | Deposit date: | 2018-08-28 | | Release date: | 2019-06-12 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and thermodynamic characterization of the binding of isoliquiritigenin to the first bromodomain of BRD4.
Febs J., 286, 2019
|
|
6AJZ
 
 | | Joint nentron and X-ray structure of BRD4 in complex with colchicin | | Descriptor: | Bromodomain-containing protein 4, N-[(7S)-1,2,3,10-tetramethoxy-9-oxo-6,7-dihydro-5H-benzo[d]heptalen-7-yl]ethanamide, SODIUM ION | | Authors: | Yokoyama, T, Ostermann, A, Schrader, T.E, Nabeshima, Y, Mizuguchi, M. | | Deposit date: | 2018-08-28 | | Release date: | 2019-06-12 | | Last modified: | 2024-03-27 | | Method: | NEUTRON DIFFRACTION (1.301 Å), X-RAY DIFFRACTION | | Cite: | Structural and thermodynamic characterization of the binding of isoliquiritigenin to the first bromodomain of BRD4.
Febs J., 286, 2019
|
|
6AJW
 
 | | Crystal structure of BRD4 in complex with DMSO (Cocktail No. 4) | | Descriptor: | Bromodomain-containing protein 4, DIMETHYL SULFOXIDE, SODIUM ION | | Authors: | Yokoyama, T, Matsumoto, K, Nabeshima, Y, Mizuguchi, M. | | Deposit date: | 2018-08-28 | | Release date: | 2019-06-12 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.401 Å) | | Cite: | Structural and thermodynamic characterization of the binding of isoliquiritigenin to the first bromodomain of BRD4.
Febs J., 286, 2019
|
|
6IMY
 
 | | Crystal structure of V30M mutated transthyretin in complex with 4'-caroboxybenzo-18-Crown-6 | | Descriptor: | 2,3,5,6,8,9,11,12,14,15-decahydro-1,4,7,10,13,16-benzohexaoxacyclooctadecine-18-carboxylic acid, Transthyretin | | Authors: | Yokoyama, T, Kosaka, Y, Matsumoto, K, Kitakami, R, Nabeshima, Y, Mizuguchi, M. | | Deposit date: | 2018-10-24 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Crown Ethers as Transthyretin Amyloidogenesis Inhibitors.
J. Med. Chem., 62, 2019
|
|
6HJ4
 
 | | Crystal structure of Whitewater Arroyo virus GP1 glycoprotein at pH 7.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CADMIUM ION, Pre-glycoprotein polyprotein GP complex | | Authors: | Pryce, R, Ng, W.M, Zeltina, A, Watanabe, Y, El Omari, K, Wagner, A, Bowden, T.A. | | Deposit date: | 2018-08-31 | | Release date: | 2018-10-10 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structure-Based Classification Defines the Discrete Conformational Classes Adopted by the Arenaviral GP1.
J. Virol., 93, 2019
|
|
