3A72
 
 | |
5JGE
 
 | |
5GV0
 
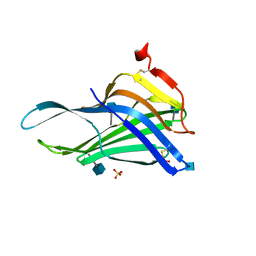 | | Crystal structure of the membrane-proximal domain of mouse lysosome-associated membrane protein 1 (LAMP-1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Lysosome-associated membrane glycoprotein 1, SULFATE ION | | Authors: | Tomabechi, Y, Ehara, H, Kukimoto-Niino, M, Shirouzu, M. | | Deposit date: | 2016-09-01 | | Release date: | 2016-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Lysosome-associated membrane proteins-1 and -2 (LAMP-1 and LAMP-2) assemble via distinct modes
Biochem.Biophys.Res.Commun., 479, 2016
|
|
7EG2
 
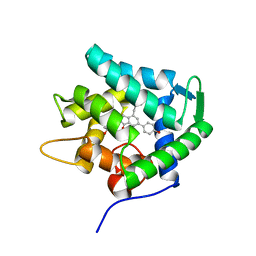 | | Crystal structure of the apoAequorin complex with (S)-daCTZ | | Descriptor: | (2~{S})-2-(hydroxymethyl)-6-(4-hydroxyphenyl)-2-[(4-hydroxyphenyl)methyl]-4-(phenylmethyl)-3~{H}-inden-1-one, Aequorin-2 | | Authors: | Tomabechi, Y, Shirouzu, M. | | Deposit date: | 2021-03-24 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Chiral deaza-coelenterazine analogs for probing a substrate-binding site in the Ca2+-binding photoprotein aequorin.
Plos One, 16, 2021
|
|
7EG3
 
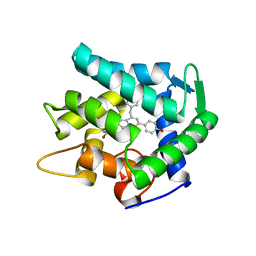 | | Crystal structure of the apoAequorin complex with (S)-HM-daCTZ | | Descriptor: | (2~{S})-6-(4-hydroxyphenyl)-2-[(4-hydroxyphenyl)methyl]-4-(phenylmethyl)-2,3-dihydroinden-1-one, Aequorin-2 | | Authors: | Tomabechi, Y, Shirouzu, M. | | Deposit date: | 2021-03-24 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Chiral deaza-coelenterazine analogs for probing a substrate-binding site in the Ca2+-binding photoprotein aequorin.
Plos One, 16, 2021
|
|
7YPD
 
 | |
5B0U
 
 | |
2KZK
 
 | | Solution structure of alpha-mannosidase binding domain of Atg34 | | Descriptor: | Uncharacterized protein YOL083W | | Authors: | Watanabe, Y, Noda, N, Kumeta, H, Suzuki, K, Ohsumi, Y, Inagaki, F. | | Deposit date: | 2010-06-18 | | Release date: | 2010-07-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Selective transport of alpha-mannosidase by autophagic pathways: structural basis for cargo recognition by Atg19 and Atg34.
J.Biol.Chem., 285, 2010
|
|
2KZB
 
 | | Solution structure of alpha-mannosidase binding domain of Atg19 | | Descriptor: | Autophagy-related protein 19 | | Authors: | Watanabe, Y, Noda, N, Kumeta, H, Suzuki, K, Ohsumi, Y, Inagaki, F. | | Deposit date: | 2010-06-15 | | Release date: | 2010-07-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Selective transport of alpha-mannosidase by autophagic pathways: structural basis for cargo recognition by Atg19 and Atg34.
J.Biol.Chem., 285, 2010
|
|
5AXD
 
 | |
5AXB
 
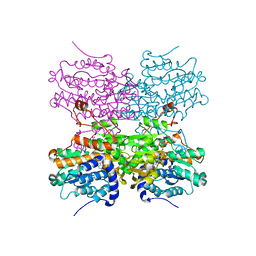 | | Crystal structure of mouse SAHH complexed with noraristeromycin | | Descriptor: | (1S,2R,3S,4R)-4-(6-aminopurin-9-yl)cyclopentane-1,2,3-triol, Adenosylhomocysteinase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Kusakabe, Y, Ishihara, M, Tanaka, N. | | Deposit date: | 2015-07-24 | | Release date: | 2016-07-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of mouse SAHH complexed with noraristeromycin
To Be Published
|
|
5AXA
 
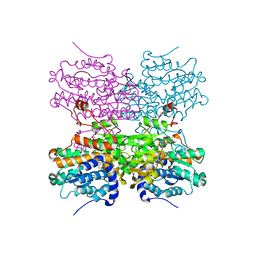 | |
5AXC
 
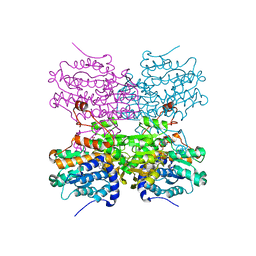 | | Crystal structure of mouse SAHH complexed with 3'-keto aristeromycin | | Descriptor: | (2S,3R,5R)-3-(6-amino-9H-purin-9-yl)-2-hydroxy-5-(hydroxymethyl)cyclopentanone, Adenosylhomocysteinase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Kusakabe, Y, Ishihara, M, Tanaka, N. | | Deposit date: | 2015-07-24 | | Release date: | 2016-07-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of mouse SAHH complexed with 3'-keto aristeromycin
To Be Published
|
|
3WZU
 
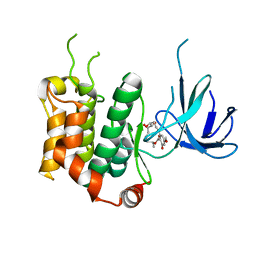 | | THE STRUCTURE OF MAP2K7 IN COMPLEX WITH 5Z-7-oxozeaenol | | Descriptor: | (3S,5Z,8S,9S,11E)-8,9,16-trihydroxy-14-methoxy-3-methyl-3,4,9,10-tetrahydro-1H-2-benzoxacyclotetradecine-1,7(8H)-dione, Dual specificity mitogen-activated protein kinase kinase 7 | | Authors: | Sogabe, Y, Hashimoto, Y, Matsumoto, T, Kinoshita, T. | | Deposit date: | 2014-10-07 | | Release date: | 2015-01-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | 5Z-7-Oxozeaenol covalently binds to MAP2K7 at Cys218 in an unprecedented manner.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
5GV3
 
 | | Crystal structure of the membrane-distal domain of mouse lysosome-associated membrane protein 2 (LAMP-2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Lysosome-associated membrane glycoprotein 2, ZINC ION | | Authors: | Tomabechi, Y, Ehara, H, Kukimoto-Niino, M, Shirouzu, M. | | Deposit date: | 2016-09-01 | | Release date: | 2017-09-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.096 Å) | | Cite: | Lysosome-associated membrane proteins-1 and -2 (LAMP-1 and LAMP-2) assemble via distinct modes.
Biochem. Biophys. Res. Commun., 479, 2016
|
|
7VSX
 
 | | Crystal structure of QL-nanoKAZ (Reverse mutant of nanoKAZ with L18Q and V27L) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, QLnK | | Authors: | Tomabechi, Y, Sekine, S, Shirouzu, M, Takamitsu, H, Satoshi, I. | | Deposit date: | 2021-10-27 | | Release date: | 2022-08-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | Reverse mutants of the catalytic 19 kDa mutant protein (nanoKAZ/nanoLuc) from Oplophorus luciferase with coelenterazine as preferred substrate.
Plos One, 17, 2022
|
|
5H0B
 
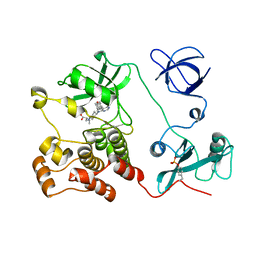 | | Crystal structure of HCK complexed with a pyrrolo-pyrimidine inhibitor (S)-2-(((1r,4S)-4-(4-amino-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-7-yl)cyclohexyl)amino)-4-methylpentanoic acid | | Descriptor: | (2~{S})-2-[[4-[4-azanyl-5-(4-phenoxyphenyl)pyrrolo[2,3-d]pyrimidin-7-yl]cyclohexyl]azaniumyl]-4-methyl-pentanoate, Tyrosine-protein kinase HCK | | Authors: | Tomabechi, Y, Kukimoto-Niino, M, Shirouzu, M. | | Deposit date: | 2016-10-04 | | Release date: | 2017-10-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | Activity cliff for 7-substituted pyrrolo-pyrimidine inhibitors of HCK explained in terms of predicted basicity of the amine nitrogen.
Bioorg. Med. Chem., 25, 2017
|
|
5H0E
 
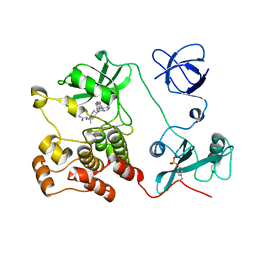 | | Crystal structure of HCK complexed with a pyrrolo-pyrimidine inhibitor (S)-2-(((1r,4S)-4-(4-amino-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-7-yl)cyclohexyl)amino)-4-methylpentanamide | | Descriptor: | (2~{S})-2-[[4-[4-azanyl-5-(4-phenoxyphenyl)pyrrolo[2,3-d]pyrimidin-7-yl]cyclohexyl]amino]-4-methyl-pentanamide, Tyrosine-protein kinase HCK | | Authors: | Tomabechi, Y, Kukimoto-Niino, M, Shirouzu, M. | | Deposit date: | 2016-10-04 | | Release date: | 2017-10-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Activity cliff for 7-substituted pyrrolo-pyrimidine inhibitors of HCK explained in terms of predicted basicity of the amine nitrogen.
Bioorg. Med. Chem., 25, 2017
|
|
5H0H
 
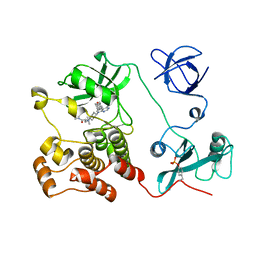 | | Crystal structure of HCK complexed with a pyrrolo-pyrimidine inhibitor (S)-2-(((1r,4S)-4-(4-amino-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-7-yl)cyclohexyl)amino)-N,N,4-trimethylpentanamide | | Descriptor: | (2~{S})-2-[[4-[4-azanyl-5-(4-phenoxyphenyl)pyrrolo[2,3-d]pyrimidin-7-yl]cyclohexyl]amino]-~{N},~{N},4-trimethyl-pentanamide, Tyrosine-protein kinase HCK | | Authors: | Tomabechi, Y, Kukimoto-Niino, M, Shirouzu, M. | | Deposit date: | 2016-10-04 | | Release date: | 2017-10-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Activity cliff for 7-substituted pyrrolo-pyrimidine inhibitors of HCK explained in terms of predicted basicity of the amine nitrogen.
Bioorg. Med. Chem., 25, 2017
|
|
5H0G
 
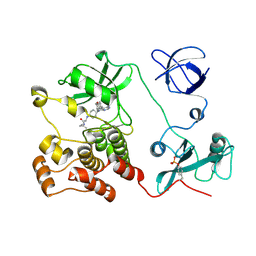 | | Crystal structure of HCK complexed with a pyrrolo-pyrimidine inhibitor (S)-2-(((1r,4S)-4-(4-amino-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-7-yl)cyclohexyl)amino)-N,4-dimethylpentanamide | | Descriptor: | (2~{S})-2-[[4-[4-azanyl-5-(4-phenoxyphenyl)pyrrolo[2,3-d]pyrimidin-7-yl]cyclohexyl]amino]-~{N},4-dimethyl-pentanamide, Tyrosine-protein kinase HCK | | Authors: | Tomabechi, Y, Kukimoto-Niino, M, Shirouzu, M. | | Deposit date: | 2016-10-04 | | Release date: | 2017-10-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Activity cliff for 7-substituted pyrrolo-pyrimidine inhibitors of HCK explained in terms of predicted basicity of the amine nitrogen.
Bioorg. Med. Chem., 25, 2017
|
|
5AUJ
 
 | |
5H09
 
 | | Crystal structure of HCK complexed with a pyrrolo-pyrimidine inhibitor (S)-ethyl2-(((1r,4S)-4-(4-amino-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-7-yl)cyclohexyl)amino)-4-methylpentanoate | | Descriptor: | Tyrosine-protein kinase HCK, ethyl (2~{S})-2-[[4-[4-azanyl-5-(4-phenoxyphenyl)pyrrolo[2,3-d]pyrimidin-7-yl]cyclohexyl]amino]-4-methyl-pentanoate | | Authors: | Tomabechi, Y, Kukimoto-Niino, M, Shirouzu, M. | | Deposit date: | 2016-10-04 | | Release date: | 2017-10-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.945 Å) | | Cite: | Activity cliff for 7-substituted pyrrolo-pyrimidine inhibitors of HCK explained in terms of predicted basicity of the amine nitrogen.
Bioorg. Med. Chem., 25, 2017
|
|
4YTV
 
 | | Crystal structure of Mdm35 | | Descriptor: | COBALT (II) ION, GLYCEROL, Mitochondrial distribution and morphology protein 35 | | Authors: | Watanabe, Y, Tamura, Y, Kawano, S, Endo, T. | | Deposit date: | 2015-03-18 | | Release date: | 2015-08-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural and mechanistic insights into phospholipid transfer by Ups1-Mdm35 in mitochondria.
Nat Commun, 6, 2015
|
|
4YTW
 
 | | Crystal structure of Ups1-Mdm35 complex | | Descriptor: | Mitochondrial distribution and morphology protein 35, Protein UPS1, mitochondrial | | Authors: | Watanabe, Y, Tamura, Y, Kawano, S, Endo, T. | | Deposit date: | 2015-03-18 | | Release date: | 2015-08-12 | | Last modified: | 2020-02-05 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural and mechanistic insights into phospholipid transfer by Ups1-Mdm35 in mitochondria.
Nat Commun, 6, 2015
|
|
4YTX
 
 | | Crystal structure of Ups1-Mdm35 complex with PA | | Descriptor: | 1,2-DILAUROYL-SN-GLYCERO-3-PHOSPHATE, Mitochondrial distribution and morphology protein 35, Protein UPS1, ... | | Authors: | Watanabe, Y, Tamura, Y, Kawano, S, Endo, T. | | Deposit date: | 2015-03-18 | | Release date: | 2015-08-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural and mechanistic insights into phospholipid transfer by Ups1-Mdm35 in mitochondria.
Nat Commun, 6, 2015
|
|
