3CQA
 
 | | Crystal structure of human fibroblast growth factor-1 with mutations Glu81Ala and Lys101Ala | | Descriptor: | FORMIC ACID, Heparin-binding growth factor 1, SULFATE ION | | Authors: | Meher, A.K, Honjo, E, Kuroki, R, Lee, J, Somasundaram, T, Blaber, M. | | Deposit date: | 2008-04-02 | | Release date: | 2009-04-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Engineering an improved crystal contact across a solvent-mediated interface of human fibroblast growth factor 1.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
1RG8
 
 | |
3CRI
 
 | | Crystal structure of human fibroblast growth factor-1 with mutations Glu81Ser, Glu82Asn and Lys101Ala | | Descriptor: | FORMIC ACID, Heparin-binding growth factor 1, SULFATE ION | | Authors: | Meher, A.K, Honjo, E, Kuroki, R, Lee, J, Somasundaram, T, Blaber, M. | | Deposit date: | 2008-04-07 | | Release date: | 2009-02-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Engineering an improved crystal contact across a solvent-mediated interface of human fibroblast growth factor 1.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
3CRH
 
 | | Crystal structure of human fibroblast growth factor-1 with mutations Glu81Ser and Lys101Ala | | Descriptor: | Heparin-binding growth factor 1, SULFATE ION | | Authors: | Meher, A.K, Honjo, E, Kuroki, R, Lee, J, Somasundaram, T, Blaber, M. | | Deposit date: | 2008-04-07 | | Release date: | 2009-02-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Engineering an improved crystal contact across a solvent-mediated interface of human fibroblast growth factor 1.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
1IT7
 
 | | Crystal structure of archaeosine tRNA-guanine transglycosylase complexed with guanine | | Descriptor: | Archaeosine tRNA-guanine transglycosylase, GUANINE, MAGNESIUM ION, ... | | Authors: | Ishitani, R, Nureki, O, Fukai, S, Kijimoto, T, Nameki, N, Watanabe, M, Kondo, H, Sekine, M, Okada, N, Nishimura, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-01-11 | | Release date: | 2002-05-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of archaeosine tRNA-guanine transglycosylase.
J.Mol.Biol., 318, 2002
|
|
1IT8
 
 | | Crystal structure of archaeosine tRNA-guanine transglycosylase from Pyrococcus horikoshii complexed with archaeosine precursor, preQ0 | | Descriptor: | 2-AMINO-4-OXO-4,7-DIHYDRO-3H-PYRROLO[2,3-D]PYRIMIDINE-5-CARBONITRILE, MAGNESIUM ION, ZINC ION, ... | | Authors: | Ishitani, R, Nureki, O, Fukai, S, Kijimoto, T, Nameki, N, Watanabe, M, Kondo, H, Sekine, M, Okada, N, Nishimura, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-01-11 | | Release date: | 2002-05-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of archaeosine tRNA-guanine transglycosylase.
J.Mol.Biol., 318, 2002
|
|
1PZZ
 
 | | Crystal structure of FGF-1, V51N mutant | | Descriptor: | FORMIC ACID, Heparin-binding growth factor 1, SULFATE ION | | Authors: | Kim, J, Blaber, M. | | Deposit date: | 2003-07-14 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Sequence swapping does not result in conformation swapping for the beta4/beta5 and beta8/beta9 beta-hairpin turns in human acidic fibroblast growth factor
Protein Sci., 14, 2005
|
|
1Q04
 
 | | Crystal structure of FGF-1, S50E/V51N | | Descriptor: | FORMIC ACID, Heparin-binding growth factor 1 | | Authors: | Kim, J, Blaber, M. | | Deposit date: | 2003-07-15 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Sequence swapping does not result in conformation swapping for the beta4/beta5 and beta8/beta9 beta-hairpin turns in human acidic fibroblast growth factor
Protein Sci., 14, 2005
|
|
1V7O
 
 | |
1Q03
 
 | | Crystal structure of FGF-1, S50G/V51G mutant | | Descriptor: | Heparin-binding growth factor 1 | | Authors: | Kim, J, Blaber, M. | | Deposit date: | 2003-07-15 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Sequence swapping does not result in conformation swapping for the beta4/beta5 and beta8/beta9 beta-hairpin turns in human acidic fibroblast growth factor
Protein Sci., 14, 2005
|
|
1P63
 
 | | Human Acidic Fibroblast Growth Factor. 140 Amino Acid Form with Amino Terminal His Tag and Leu111 Replaced with Ile (L111I) | | Descriptor: | ACIDIC FIBROBLAST GROWTH FACTOR, FORMIC ACID, SULFATE ION | | Authors: | Brych, S.R, Kim, J, Logan, T.M, Blaber, M. | | Deposit date: | 2003-04-28 | | Release date: | 2004-05-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Accommodation of a highly symmetric core within a symmetric protein
superfold
Protein Sci., 12, 2003
|
|
1ZBJ
 
 | |
1JY0
 
 | |
5ABS
 
 | | CRYSTAL STRUCTURE OF THE C-TERMINAL COILED-COIL DOMAIN OF CIN85 IN SPACE GROUP P321 | | Descriptor: | SH3 DOMAIN-CONTAINING KINASE-BINDING PROTEIN 1, ZINC ION | | Authors: | Wong, L, Habeck, M, Griesinger, C, Becker, S. | | Deposit date: | 2015-08-07 | | Release date: | 2016-07-13 | | Last modified: | 2019-02-06 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | The Adaptor Protein Cin85 Assembles Intracellular Signaling Clusters for B Cell Activation.
Sci.Signal., 9, 2016
|
|
1Z2V
 
 | |
1Z4S
 
 | |
5AX2
 
 | | Crystal structure of S.cerevisiae Kti11p | | Descriptor: | CADMIUM ION, Diphthamide biosynthesis protein 3 | | Authors: | Kumar, A, Nagarathinam, K, Tanabe, M, Balbach, J. | | Deposit date: | 2015-07-13 | | Release date: | 2016-07-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Hyperbolic Pressure-Temperature Phase Diagram of the Zinc-Finger Protein apoKti11 Detected by NMR Spectroscopy.
J Phys Chem B, 123, 2019
|
|
5B5L
 
 | | Crystal structure of acetyl esterase mutant S10A with acetate ion | | Descriptor: | ACETATE ION, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Uechi, K, Kamachi, S, Akita, H, Mine, S, Watanabe, M. | | Deposit date: | 2016-05-12 | | Release date: | 2017-05-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | crystal structure of acetyl esterase mutant S10A with acetate ion
To Be Published
|
|
1M9H
 
 | | Corynebacterium 2,5-DKGR A and Phe 22 replaced with Tyr (F22Y), Lys 232 replaced with Gly (K232G), Arg 238 replaced with His (R238H)and Ala 272 replaced with Gly (A272G)in presence of NADH cofactor | | Descriptor: | 2,5-diketo-D-gluconic acid reductase A, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | Authors: | Sanli, G, Blaber, M. | | Deposit date: | 2002-07-29 | | Release date: | 2003-08-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural alteration of cofactor specificity in Corynebacterium 2,5-diketo-D-gluconic acid reductase
Protein Sci., 13, 2004
|
|
3AQD
 
 | |
2YU0
 
 | | Solution structures of the PAAD_DAPIN domain of mus musculus interferon-activatable protein 205 | | Descriptor: | Interferon-activable protein 205 | | Authors: | Sato, M, Tochio, N, Koshiba, S, Watanabe, M, Harada, T, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-05 | | Release date: | 2008-02-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the PAAD_DAPIN domain of mus musculus interferon-activatable protein 205
To be Published
|
|
1V9Y
 
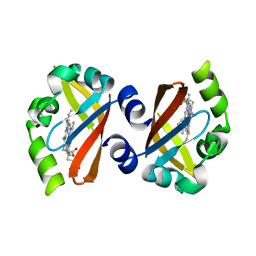 | | Crystal Structure of the heme PAS sensor domain of Ec DOS (ferric form) | | Descriptor: | Heme pas sensor protein, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Kurokawa, H, Lee, D.S, Watanabe, M, Sagami, I, Mikami, B, Raman, C.S, Shimizu, T. | | Deposit date: | 2004-02-04 | | Release date: | 2004-05-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | A redox-controlled molecular switch revealed by the crystal structure of a bacterial heme PAS sensor.
J.Biol.Chem., 279, 2004
|
|
3AK0
 
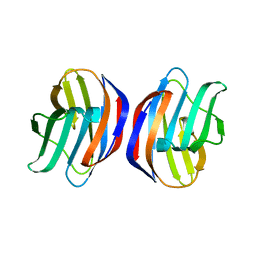 | | Crystal Structure of Ancestral Congerin Con-anc'-N28K | | Descriptor: | Ancestral congerin Con-anc, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Konno, A, Kitagawa, A, Watanabe, M, Ogawa, T, Shirai, T. | | Deposit date: | 2010-06-29 | | Release date: | 2011-05-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Tracing protein evolution through ancestral structures of fish galectin
Structure, 19, 2011
|
|
3AJY
 
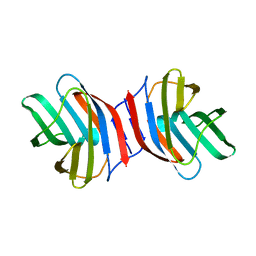 | | Crystal Structure of Ancestral Congerin Con-anc | | Descriptor: | Ancestral congerin Con-anc, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Konno, A, Kitagawa, A, Watanabe, M, Ogawa, T, Shirai, T. | | Deposit date: | 2010-06-29 | | Release date: | 2011-05-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Tracing protein evolution through ancestral structures of fish galectin
Structure, 19, 2011
|
|
1V9Z
 
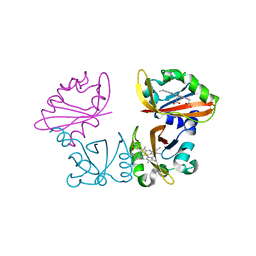 | | Crystal Structure of the heme PAS sensor domain of Ec DOS (Ferrous Form) | | Descriptor: | Heme pas sensor protein, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Kurokawa, H, Lee, D.S, Watanabe, M, Sagami, I, Mikami, B, Raman, C.S, Shimizu, T. | | Deposit date: | 2004-02-04 | | Release date: | 2004-05-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A redox-controlled molecular switch revealed by the crystal structure of a bacterial heme PAS sensor.
J.Biol.Chem., 279, 2004
|
|
