3AJZ
 
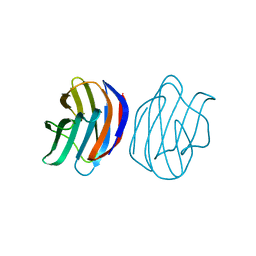 | | Crystal Structure of Ancestral Congerin Con-anc | | Descriptor: | Ancestral congerin Con-anc, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Konno, A, Kitagawa, A, Watanabe, M, Ogawa, T, Shirai, T. | | Deposit date: | 2010-06-29 | | Release date: | 2011-05-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Tracing protein evolution through ancestral structures of fish galectin
Structure, 19, 2011
|
|
1VB6
 
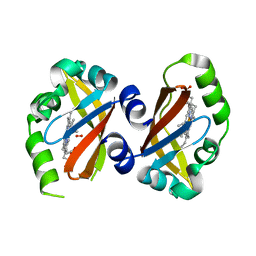 | | Crystal Structure of the heme PAS sensor domain of Ec DOS (oxygen-bound form) | | Descriptor: | Heme pas sensor protein, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Kurokawa, H, Watanabe, M, Sagami, I, Mikami, B, Shimizu, T. | | Deposit date: | 2004-02-24 | | Release date: | 2005-04-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structure of oxygen-bound form of a Heme PAS domain of Ec DOS
To be Published
|
|
3OYP
 
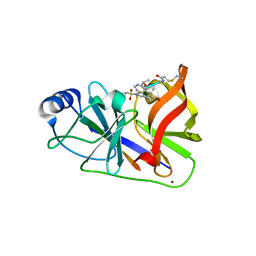 | | HCV NS3/4A in complex with ligand 3 | | Descriptor: | Non-structural protein 4A, Peptidomimetic inhibitor, Serine protease NS3, ... | | Authors: | Hagel, M, Niu, D, St.Martin, T, Sheets, M.P, Qiao, L, Bernard, H, Karp, R.M, Zhu, Z, Labenski, M.T, Chaturvedi, P.C, Nacht, M, Westlin, W.F, Petter, R.C, Singh, J. | | Deposit date: | 2010-09-23 | | Release date: | 2010-12-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Selective irreversible inhibition of a protease by targeting a noncatalytic cysteine.
Nat.Chem.Biol., 7, 2011
|
|
3RQ7
 
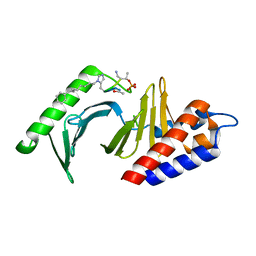 | | Polo-like kinase 1 Polo box domain in complex with a C6H5(CH2)8-derivatized peptide inhibitor | | Descriptor: | C6H5(CH2)8-derivatized peptide inhibitor, Serine/threonine-protein kinase PLK1 | | Authors: | Liu, F, Park, J.-E, Qian, W.-J, Lim, D.C, Graber, M, Berg, T, Yaffe, M.B, Lee, K.S, Burke Jr, T.R. | | Deposit date: | 2011-04-27 | | Release date: | 2011-07-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Serendipitous alkylation of a Plk1 ligand uncovers a new binding channel.
Nat.Chem.Biol., 7, 2011
|
|
6KA4
 
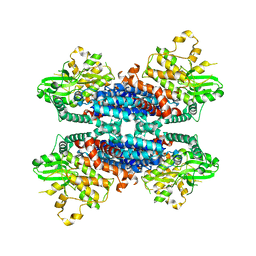 | | Cryo-EM structure of the AtMLKL3 tetramer | | Descriptor: | F22L4.1 protein | | Authors: | Lisa, M, Huang, M, Zhang, X, Ryohei, T.N, Leila, B.K, Isabel, M.L.S, Florence, J, Viera, K, Dmitry, L, Jane, E.P, James, M.M, Kay, H, Paul, S.L, Chai, J, Takaki, M. | | Deposit date: | 2019-06-20 | | Release date: | 2020-09-23 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure of the AtMLKL3 tetramer
To Be Published
|
|
2CZW
 
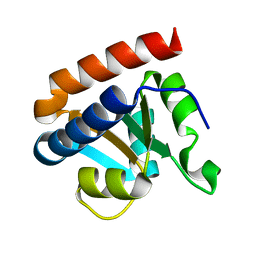 | | Crystal structure analysis of protein component Ph1496p of P.horikoshii ribonuclease P | | Descriptor: | 50S ribosomal protein L7Ae | | Authors: | Fukuhara, H, Kifusa, M, Watanabe, M, Terada, A, Honda, T, Numata, T, Kakuta, Y, Kimura, M. | | Deposit date: | 2005-07-19 | | Release date: | 2006-04-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A fifth protein subunit Ph1496p elevates the optimum temperature for the ribonuclease P activity from Pyrococcus horikoshii OT3
Biochem.Biophys.Res.Commun., 343, 2006
|
|
2DVY
 
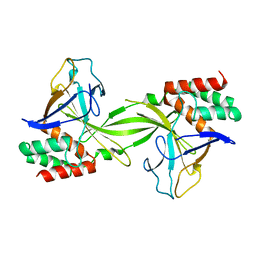 | | Crystal structure of restriction endonucleases PabI | | Descriptor: | Restriction endonuclease PabI | | Authors: | Miyazono, K, Watanabe, M, Kamo, M, Sawasaki, T, Nagata, K, Endo, Y, Tanokura, M, Kobayashi, I. | | Deposit date: | 2006-08-01 | | Release date: | 2007-05-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Novel protein fold discovered in the PabI family of restriction enzymes
Nucleic Acids Res., 35, 2007
|
|
5XMH
 
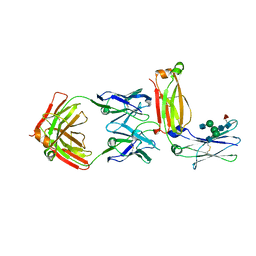 | | Crystal structure of an IgM rheumatoid factor YES8c in complex with IgG1 Fc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Immunoglobulin gamma-1 heavy chain, ... | | Authors: | Shiroishi, M, Shimokawa, K, Lee, J.M, Kusakabe, M, Ueda, T. | | Deposit date: | 2017-05-15 | | Release date: | 2018-03-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-function analyses of a stereotypic rheumatoid factor unravel the structural basis for germline-encoded antibody autoreactivity.
J. Biol. Chem., 293, 2018
|
|
6KFQ
 
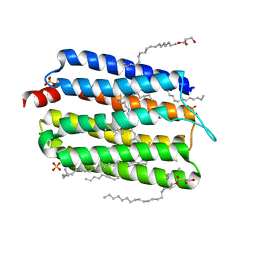 | | Crystal structure of thermophilic rhodopsin from Rubrobacter xylanophilus | | Descriptor: | RETINAL, Rhodopsin, SULFATE ION, ... | | Authors: | Suzuki, K, Akiyama, T, Hayashi, T, Yasuda, S, Kanehara, K, Kojima, K, Tanabe, M, Kato, R, Senda, T, Sudo, Y, Kinoshita, M, Murata, T. | | Deposit date: | 2019-07-08 | | Release date: | 2020-04-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | How Does a Microbial Rhodopsin RxR Realize Its Exceptionally High Thermostability with the Proton-Pumping Function Being Retained?
J.Phys.Chem.B, 124, 2020
|
|
7COQ
 
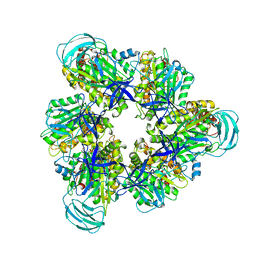 | |
3O3Q
 
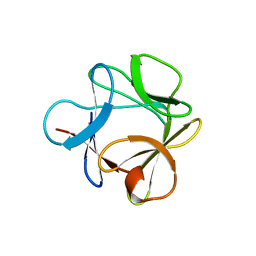 | |
3O49
 
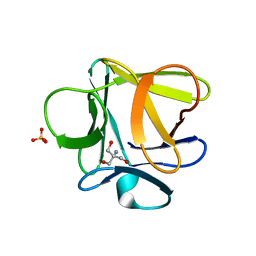 | |
3OL0
 
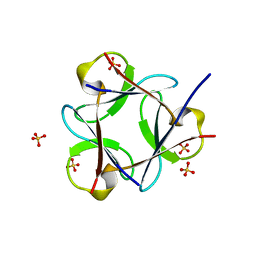 | |
3OGF
 
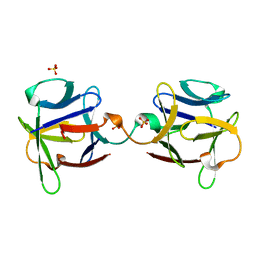 | |
2NTD
 
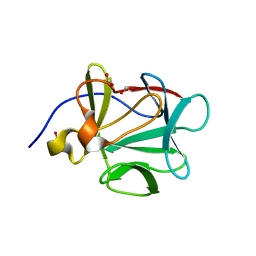 | |
2KWC
 
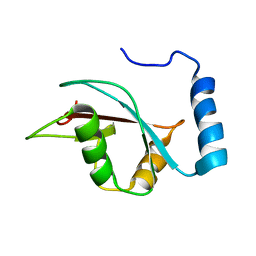 | | The NMR structure of the autophagy-related protein Atg8 | | Descriptor: | Autophagy-related protein 8 | | Authors: | Kumeta, H, Watanabe, M, Nakatogawa, H, Yamaguchi, M, Ogura, K, Adachi, W, Fujioka, Y, Noda, N.N, Ohsumi, Y, Inagaki, F. | | Deposit date: | 2010-04-05 | | Release date: | 2010-05-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The NMR structure of the autophagy-related protein Atg8
J.Biomol.Nmr, 47, 2010
|
|
2LME
 
 | | Solid-state NMR structure of the membrane anchor domain of the trimeric autotransporter YadA | | Descriptor: | Adhesin yadA | | Authors: | Shahid, S.A, Bardiaux, B, Franks, W.T, Habeck, M, Linke, D, van Rossum, B. | | Deposit date: | 2011-11-30 | | Release date: | 2012-11-07 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Membrane-protein structure determination by solid-state NMR spectroscopy of microcrystals.
Nat.Methods, 9, 2012
|
|
2MF4
 
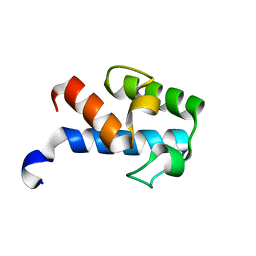 | | 1H, 13C, 15N chemical shift assignments of Streptomyces virginiae VirA acp5a | | Descriptor: | Hybrid polyketide synthase-non ribosomal peptide synthetase | | Authors: | Davison, J, Dorival, J, Rabeharindranto, M.H, Mazon, H, Chagot, B, Gruez, A, Weissman, K.J. | | Deposit date: | 2013-10-05 | | Release date: | 2014-06-04 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR assignements of ACP5a
To be Published
|
|
3VTE
 
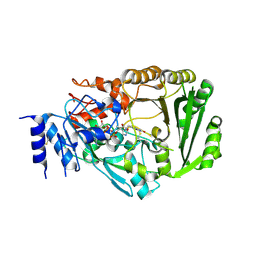 | | Crystal structure of tetrahydrocannabinolic acid synthase from Cannabis sativa | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, Tetrahydrocannabinolic acid synthase | | Authors: | Shoyama, Y, Tamada, T, Kurihara, K, Takeuchi, A, Taura, F, Arai, S, Blaber, M, Shoyama, Y, Morimoto, S, Kuroki, R. | | Deposit date: | 2012-05-28 | | Release date: | 2012-07-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure and function of 1-tetrahydrocannabinolic acid (THCA) synthase, the enzyme controlling the psychoactivity of Cannabis sativa
J.Mol.Biol., 423, 2012
|
|
3WI5
 
 | | Crystal structure of the Loop 7 mutant PorB from Neisseria meningitidis serogroup B | | Descriptor: | CITRATE ANION, Major outer membrane protein P.IB | | Authors: | Kattner, C, Toussi, D, Wetzler, L.M, Ruppel, N, Massari, P, Tanabe, M. | | Deposit date: | 2013-09-05 | | Release date: | 2014-01-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystallographic analysis of Neisseria meningitidis PorB extracellular loops potentially implicated in TLR2 recognition.
J.Struct.Biol., 185, 2014
|
|
3W2T
 
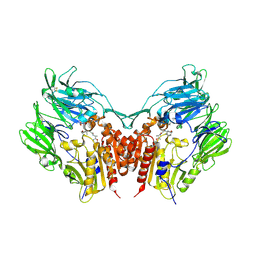 | | Crystal structure of human depiptidyl peptidase IV (DPP-4) in complex with vildagliptin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-{[(1r,3s,5R,7S)-3-hydroxytricyclo[3.3.1.1~3,7~]decan-1-yl]amino}-1-{(2S)-2-[(E)-iminomethyl]pyrrolidin-1-yl}ethan-1-o ne, ... | | Authors: | Kishida, H, Nabeno, M, Miyaguchi, I, Tanaka, Y, Katou, R, Akahoshi, F. | | Deposit date: | 2012-12-04 | | Release date: | 2013-05-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | A comparative study of the binding modes of recently launched dipeptidyl peptidase IV inhibitors in the active site
Biochem.Biophys.Res.Commun., 434, 2013
|
|
3WI4
 
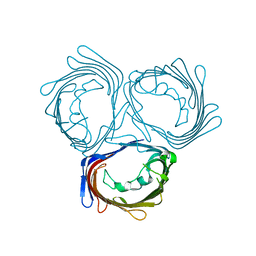 | | Crystal structure of wild-type PorB from Neisseria meningitidis serogroup B | | Descriptor: | Major outer membrane protein P.IB | | Authors: | Kattner, C, Toussi, D, Wetzler, L.M, Ruppel, N, Massari, P, Tanabe, M. | | Deposit date: | 2013-09-05 | | Release date: | 2014-01-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.32 Å) | | Cite: | Crystallographic analysis of Neisseria meningitidis PorB extracellular loops potentially implicated in TLR2 recognition.
J.Struct.Biol., 185, 2014
|
|
3VY8
 
 | | Crystal Structure of PorB from Neisseria meningitidis in complex with Cesium ion, space group P63 | | Descriptor: | CESIUM ION, Outer membrane protein | | Authors: | Kattner, C, Zaucha, J, Jaenecke, F, Zachariae, U, Tanabe, M. | | Deposit date: | 2012-09-21 | | Release date: | 2013-01-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Identification of a cation transport pathway in Neisseria meningitidis PorB.
Proteins, 81, 2013
|
|
3VY9
 
 | | Crystal structure of PorB from Neisseria meningitidis in complex with cesium ion, space group H32 | | Descriptor: | CESIUM ION, Outer membrane protein | | Authors: | Kattner, C, Zaucha, J, Jaenecke, F, Zachariae, U, Tanabe, M. | | Deposit date: | 2012-09-21 | | Release date: | 2013-01-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Identification of a cation transport pathway in Neisseria meningitidis PorB.
Proteins, 81, 2013
|
|
3AUP
 
 | | Crystal structure of Basic 7S globulin from soybean | | Descriptor: | Basic 7S globulin | | Authors: | Yoshizawa, T, Shimizu, T, Taichi, M, Nishiuchi, Y, Yamabe, M, Shichijo, N, Unzai, S, Hirano, H, Sato, M, Hashimoto, H. | | Deposit date: | 2011-02-14 | | Release date: | 2011-04-27 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Crystal structure of basic 7S globulin, a xyloglucan-specific endo-beta-1,4-glucanase inhibitor protein-like protein from soybean lacking inhibitory activity against endo-beta-glucanase
Febs J., 278, 2011
|
|
