5X9V
 
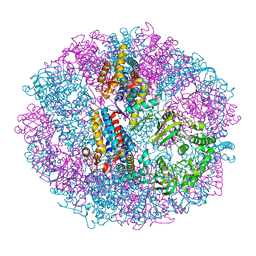 | | Crystal structure of group III chaperonin in the Closed state | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Thermosome, ... | | Authors: | An, Y.J, Cha, S.S. | | Deposit date: | 2017-03-09 | | Release date: | 2017-10-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.003 Å) | | Cite: | Structural and mechanistic characterization of an archaeal-like chaperonin from a thermophilic bacterium
Nat Commun, 8, 2017
|
|
5K1D
 
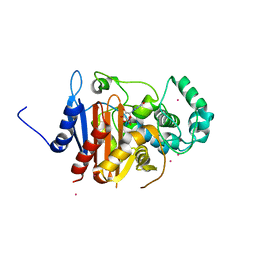 | | Crystal structure of a class C beta lactamase/compound1 complex | | Descriptor: | Beta-lactamase, CADMIUM ION, GUANOSINE-5'-MONOPHOSPHATE | | Authors: | AN, Y.J, Na, J.H, Cha, S.S. | | Deposit date: | 2016-05-18 | | Release date: | 2017-05-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | GMP and IMP Are Competitive Inhibitors of CMY-10, an Extended-Spectrum Class C beta-Lactamase.
Antimicrob. Agents Chemother., 61, 2017
|
|
5K1F
 
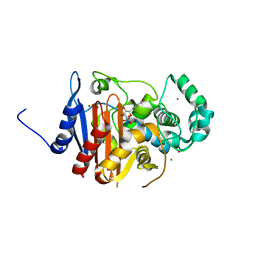 | | Crystal structure of a class C beta lactamase/compound2 complex | | Descriptor: | Beta-lactamase, CADMIUM ION, INOSINIC ACID | | Authors: | An, Y.J, Na, J.H, Cha, S.S. | | Deposit date: | 2016-05-18 | | Release date: | 2017-05-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | GMP and IMP Are Competitive Inhibitors of CMY-10, an Extended-Spectrum Class C beta-Lactamase.
Antimicrob. Agents Chemother., 61, 2017
|
|
4QF5
 
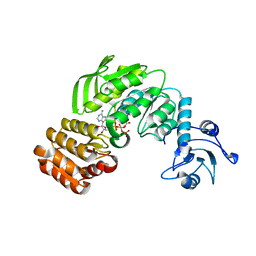 | | Crystal structure I of MurF from Acinetobacter baumannii | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, UDP-N-acetylmuramoyl-tripeptide--D-alanyl-D-alanine ligase | | Authors: | An, Y.J, Jeong, C.S, Cha, S.S. | | Deposit date: | 2014-05-19 | | Release date: | 2015-04-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | ATP-binding mode including a carbamoylated lysine and two Mg(2+) ions, and substrate-binding mode in Acinetobacter baumannii MurF
Biochem.Biophys.Res.Commun., 450, 2014
|
|
4IVI
 
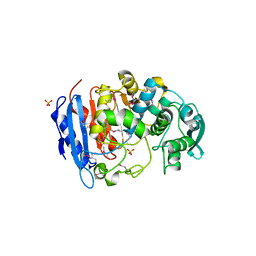 | | Crystal structure of a family VIII carboxylesterase. | | Descriptor: | Carboxylesterase, SULFATE ION | | Authors: | An, Y.J, Kim, M.-K, Jeong, C.-S, Cha, S.-S. | | Deposit date: | 2013-01-23 | | Release date: | 2013-06-19 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the beta-lactamase activity of EstU1, a family VIII carboxylesterase.
Proteins, 81, 2013
|
|
4ZPX
 
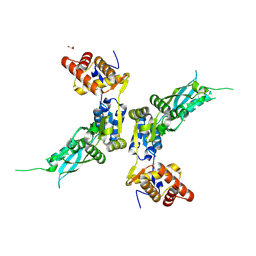 | |
4IVK
 
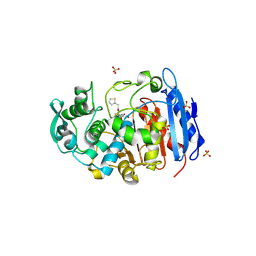 | | Crystal structure of a fammily VIII carboxylesterase in a complex with cephalothin. | | Descriptor: | CEPHALOTHIN GROUP, Carboxylesterases, SULFATE ION | | Authors: | An, Y.J, Kim, M.-K, Jeong, C.-S, Cha, S.-S. | | Deposit date: | 2013-01-23 | | Release date: | 2013-06-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the beta-lactamase activity of EstU1, a family VIII carboxylesterase.
Proteins, 81, 2013
|
|
5F1G
 
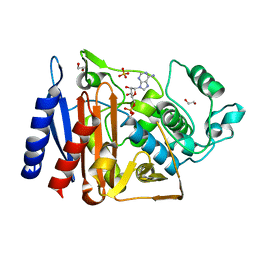 | | Crystal structure of AmpC BER adenylylated in the cytoplasm | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE MONOPHOSPHATE, Beta-lactamase, ... | | Authors: | An, Y.J, Kim, M.K, Na, J.H, Cha, S.S. | | Deposit date: | 2015-11-30 | | Release date: | 2016-12-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural and mechanistic insights into the inhibition of class C beta-lactamases through the adenylylation of the nucleophilic serine.
J.Antimicrob.Chemother., 72, 2017
|
|
5F1F
 
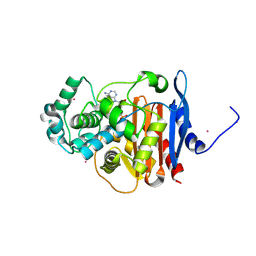 | | Crystal structure of CMY-10 adenylylated by acetyl-AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Beta-lactamase, CADMIUM ION | | Authors: | An, Y.J, Kim, M.K, Na, J.H, Cha, S.S. | | Deposit date: | 2015-11-30 | | Release date: | 2016-12-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.548 Å) | | Cite: | Structural and mechanistic insights into the inhibition of class C beta-lactamases through the adenylylation of the nucleophilic serine.
J.Antimicrob.Chemother., 72, 2017
|
|
4DT3
 
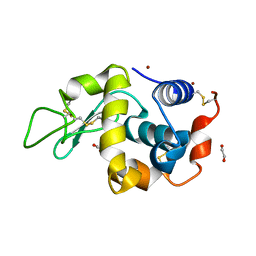 | | Crystal structure of zinc-charged lysozyme | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C, ... | | Authors: | An, Y.J, Jeong, C.S, Cha, S.S. | | Deposit date: | 2012-02-20 | | Release date: | 2012-09-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Experimental phasing using zinc anomalous scattering
Acta Crystallogr.,Sect.D, 68, 2012
|
|
5X9U
 
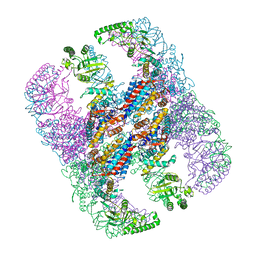 | | Crystal structure of group III chaperonin in the open state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Thermosome, alpha subunit | | Authors: | An, Y.J, Cha, S.S. | | Deposit date: | 2017-03-09 | | Release date: | 2017-10-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (4.001 Å) | | Cite: | Structural and mechanistic characterization of an archaeal-like chaperonin from a thermophilic bacterium
Nat Commun, 8, 2017
|
|
4QDI
 
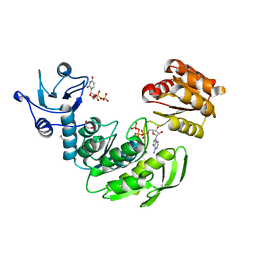 | | Crystal structure II of MurF from Acinetobacter baumannii | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | An, Y.J, Jeong, C.S, Cha, S.S. | | Deposit date: | 2014-05-13 | | Release date: | 2015-04-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | ATP-binding mode including a carbamoylated lysine and two Mg(2+) ions, and substrate-binding mode in Acinetobacter baumannii MurF
Biochem.Biophys.Res.Commun., 450, 2014
|
|
3BP8
 
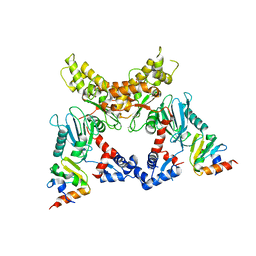 | | Crystal structure of Mlc/EIIB complex | | Descriptor: | ACETATE ION, PTS system glucose-specific EIICB component, Putative NAGC-like transcriptional regulator, ... | | Authors: | An, Y.J, Jung, H.I, Cha, S.S. | | Deposit date: | 2007-12-18 | | Release date: | 2008-05-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Analyses of Mlc-IIBGlc interaction and a plausible molecular mechanism of Mlc inactivation by membrane sequestration
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
5H4U
 
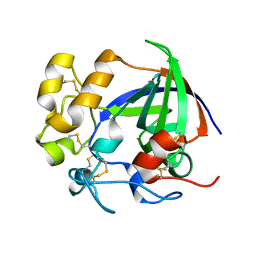 | | Crystal structure of cellulase from Antarctic springtail, Cryptopygus antarcticus | | Descriptor: | Endo-beta-1,4-glucanase | | Authors: | An, Y.J, Hong, S.K, Cha, S.S. | | Deposit date: | 2016-11-02 | | Release date: | 2017-03-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Genetic and Structural Characterization of a Thermo-Tolerant, Cold-Active, and Acidic Endo-beta-1,4-glucanase from Antarctic Springtail, Cryptopygus antarcticus.
J. Agric. Food Chem., 65, 2017
|
|
5GSC
 
 | |
5GZW
 
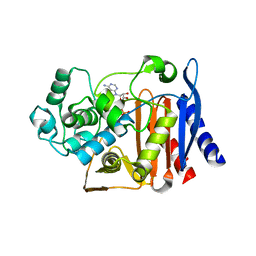 | | Crystal structure of AmpC BER adenylylated by acetyl-AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Beta-lactamase, SULFATE ION | | Authors: | An, Y.J, Cha, S.S. | | Deposit date: | 2016-10-02 | | Release date: | 2017-10-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.489 Å) | | Cite: | Structural and mechanistic insights into the inhibition of class C beta-lactamases through the adenylylation of the nucleophilic serine.
J.Antimicrob.Chemother., 72, 2017
|
|
5GGW
 
 | | Crystal structure of Class C beta-lactamase | | Descriptor: | Beta-lactamase, PHOSPHATE ION, SULFATE ION | | Authors: | An, Y.J, Na, J.H, Cha, S.S. | | Deposit date: | 2016-06-16 | | Release date: | 2017-05-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.762 Å) | | Cite: | Structural basis for the extended substrate spectrum of AmpC BER and structure-guided discovery of the inhibition activity of citrate against the class C beta-lactamases AmpC BER and CMY-10.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5GMX
 
 | | Crystal structure of a family VIII carboxylesterase | | Descriptor: | Carboxylesterase, phenylmethanesulfonic acid | | Authors: | An, Y.J, Cha, S.S. | | Deposit date: | 2016-07-18 | | Release date: | 2017-07-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of EstSRT1, a family VIII carboxylesterase displaying hydrolytic activity toward oxyimino cephalosporins
Biochem. Biophys. Res. Commun., 478, 2016
|
|
4MB0
 
 | | Crystal structure of TON1374 | | Descriptor: | ACETATE ION, phosphopantothenate synthetase | | Authors: | Kim, M.-K, An, Y.J, Cha, S.-S. | | Deposit date: | 2013-08-19 | | Release date: | 2014-08-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | The crystal structure of a novel phosphopantothenate synthetase from the hyperthermophilic archaea, Thermococcus onnurineus NA1
Biochem.Biophys.Res.Commun., 439, 2013
|
|
3K1J
 
 | | Crystal structure of Lon protease from Thermococcus onnurineus NA1 | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Cha, S.S, An, Y.J. | | Deposit date: | 2009-09-28 | | Release date: | 2010-09-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Lon protease: molecular architecture of gated entry to a sequestered degradation chamber
Embo J., 29, 2010
|
|
3MWM
 
 | |
5JOC
 
 | | Crystal structure of the S61A mutant of AmpC BER | | Descriptor: | Beta-lactamase, CITRIC ACID | | Authors: | Na, J.H, An, Y.J, Cha, S.S. | | Deposit date: | 2016-05-02 | | Release date: | 2017-05-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for the extended substrate spectrum of AmpC BER and structure-guided discovery of the inhibition activity of citrate against the class C beta-lactamases AmpC BER and CMY-10.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
3MVE
 
 | | Crystal structure of a novel pyruvate decarboxylase | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, UPF0255 protein VV1_0328 | | Authors: | Cha, S.S, Jeong, C.S, An, Y.J. | | Deposit date: | 2010-05-04 | | Release date: | 2011-05-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | FrsA functions as a cofactor-independent decarboxylase to control metabolic flux.
Nat.Chem.Biol., 7, 2011
|
|
3EYY
 
 | | Structural basis for the specialization of Nur, a nickel-specific Fur homologue, in metal sensing and DNA recognition | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, MALONATE ION, ... | | Authors: | Cha, S.-S, An, Y.J. | | Deposit date: | 2008-10-22 | | Release date: | 2009-06-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for the specialization of Nur, a nickel-specific Fur homolog, in metal sensing and DNA recognition
Nucleic Acids Res., 37, 2009
|
|
4OOU
 
 | | Crystal structure of beta-1,4-D-mannanase from Cryptopygus antarcticus | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-1,4-mannanase | | Authors: | Kim, M.-K, An, Y.J, Jeong, C.-S, Cha, S.-S. | | Deposit date: | 2014-02-04 | | Release date: | 2014-08-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structure-based investigation into the functional roles of the extended loop and substrate-recognition sites in an endo-beta-1,4-d-mannanase from the Antarctic springtail, Cryptopygus antarcticus.
Proteins, 82, 2014
|
|
