3ZPP
 
 | |
6DZN
 
 | | Pan-ebolavirus human antibody ADI-15878 Fab | | Descriptor: | 2-(2-ETHOXYETHOXY)ETHANOL, Antibody ADI-15878, heavy chain, ... | | Authors: | Murin, C.D, Ward, A.B, Bruhn, J.F, Stanfield, R. | | Deposit date: | 2018-07-05 | | Release date: | 2018-09-12 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis of Pan-Ebolavirus Neutralization by an Antibody Targeting the Glycoprotein Fusion Loop.
Cell Rep, 24, 2018
|
|
1U8E
 
 | | HUMAN DIPEPTIDYL PEPTIDASE IV/CD26 MUTANT Y547F | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bjelke, J.R, Christensen, J, Branner, S, Wagtmann, N, Olsen, C, Kanstrup, A.B, Rasmussen, H.B. | | Deposit date: | 2004-08-05 | | Release date: | 2004-08-17 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Tyrosine 547 constitutes an essential part of the catalytic mechanism of dipeptidyl peptidase IV
J.Biol.Chem., 279, 2004
|
|
1TT3
 
 | | NMR soulution structure of omega-conotoxin [K10]MVIIA | | Descriptor: | Omega-conotoxin MVIIa | | Authors: | Adams, D.J, Smith, A.B, Schroeder, C.I, Yasuda, T, Lewis, R.J. | | Deposit date: | 2004-06-21 | | Release date: | 2004-07-06 | | Last modified: | 2021-11-10 | | Method: | SOLUTION NMR | | Cite: | omega-conotoxin CVID inhibits a pharmacologically distinct voltage-sensitive calcium channel associated with transmitter release from preganglionic nerve terminals
J.Biol.Chem., 278, 2003
|
|
1TU6
 
 | | Cathepsin K complexed with a ketoamide inhibitor | | Descriptor: | Cathepsin K, SULFATE ION, [1-(4-FLUOROBENZYL)CYCLOBUTYL]METHYL (1S)-1-[OXO(1H-PYRAZOL-5-YLAMINO)ACETYL]PENTYLCARBAMATE | | Authors: | Barrett, D.G, Catalano, J.G, Deaton, D.N, Hassell, A.M, Long, S.T, Miller, A.B, Miller, L.R, Shewchuk, L.M, Wells-Knecht, K.J, Wright, L.L. | | Deposit date: | 2004-06-24 | | Release date: | 2004-09-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Potent and selective P2-P3 ketoamide inhibitors of cathepsin K with improved pharmacokinetic properties via favorable P1', P1, and/or P3 substitutions
BIOORG.MED.CHEM.LETT., 14, 2004
|
|
1UB6
 
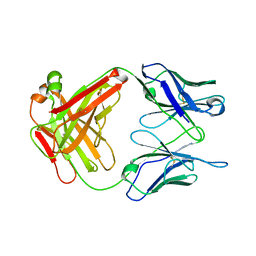 | | Crystal structure of Antibody 19G2 with sera ligand | | Descriptor: | antibody 19G2, alpha chain, beta chain | | Authors: | Beuscher, A.B, Wirsching, P, Lerner, R.A, Janda, K, Stevens, R.C. | | Deposit date: | 2003-03-30 | | Release date: | 2004-04-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structure and Dynamics of Blue Fluorescent Antibody 19G2 at Blue and Violet Fluorescent Temperatures
To be published
|
|
1KNL
 
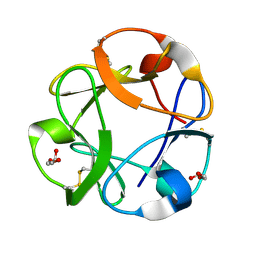 | | Streptomyces lividans Xylan Binding Domain cbm13 | | Descriptor: | ENDO-1,4-BETA-XYLANASE A, GLYCEROL | | Authors: | Notenboom, V, Boraston, A.B, Williams, S.J, Kilburn, D.G, Rose, D.R. | | Deposit date: | 2001-12-19 | | Release date: | 2002-06-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | High-resolution crystal structures of the lectin-like xylan binding domain from Streptomyces lividans xylanase 10A with bound substrates reveal a novel mode of xylan binding.
Biochemistry, 41, 2002
|
|
1KLA
 
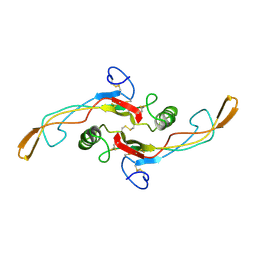 | | SOLUTION STRUCTURE OF TGF-B1, NMR, MODELS 1-17 OF 33 STRUCTURES | | Descriptor: | TRANSFORMING GROWTH FACTOR-BETA 1 | | Authors: | Hinck, A.P, Archer, S.J, Qian, S.W, Roberts, A.B, Sporn, M.B, Weatherbee, J.A, Tsang, M.L.-S, Lucas, R, Zhang, B.-L, Wenker, J, Torchia, D.A. | | Deposit date: | 1996-01-16 | | Release date: | 1996-08-17 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Transforming growth factor beta 1: three-dimensional structure in solution and comparison with the X-ray structure of transforming growth factor beta 2.
Biochemistry, 35, 1996
|
|
1KNM
 
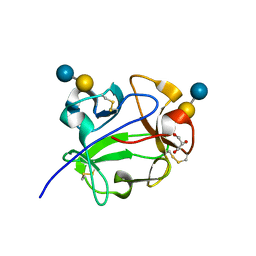 | | Streptomyces lividans Xylan Binding Domain cbm13 in Complex with Lactose | | Descriptor: | ENDO-1,4-BETA-XYLANASE A, GLYCEROL, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Notenboom, V, Boraston, A.B, Williams, S.J, Kilburn, D.G, Rose, D.R. | | Deposit date: | 2001-12-19 | | Release date: | 2002-06-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | High-resolution crystal structures of the lectin-like xylan binding domain from Streptomyces lividans xylanase 10A with bound substrates reveal a novel mode of xylan binding.
Biochemistry, 41, 2002
|
|
1KLC
 
 | | SOLUTION STRUCTURE OF TGF-B1, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | TRANSFORMING GROWTH FACTOR-BETA 1 | | Authors: | Hinck, A.P, Archer, S.J, Qian, S.W, Roberts, A.B, Sporn, M.B, Weatherbee, J.A, Tsang, M.L.-S, Lucas, R, Zhang, B.-L, Wenker, J, Torchia, D.A. | | Deposit date: | 1996-01-16 | | Release date: | 1996-08-17 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Transforming growth factor beta 1: three-dimensional structure in solution and comparison with the X-ray structure of transforming growth factor beta 2.
Biochemistry, 35, 1996
|
|
1KLD
 
 | | SOLUTION STRUCTURE OF TGF-B1, NMR, MODELS 18-33 OF 33 STRUCTURES | | Descriptor: | TRANSFORMING GROWTH FACTOR-BETA 1 | | Authors: | Hinck, A.P, Archer, S.J, Qian, S.W, Roberts, A.B, Sporn, M.B, Weatherbee, J.A, Tsang, M.L.-S, Lucas, R, Zhang, B.-L, Wenker, J, Torchia, D.A. | | Deposit date: | 1996-01-16 | | Release date: | 1996-08-17 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Transforming growth factor beta 1: three-dimensional structure in solution and comparison with the X-ray structure of transforming growth factor beta 2.
Biochemistry, 35, 1996
|
|
1KTZ
 
 | | Crystal Structure of the Human TGF-beta Type II Receptor Extracellular Domain in Complex with TGF-beta3 | | Descriptor: | TGF-beta Type II Receptor, TRANSFORMING GROWTH FACTOR BETA 3 | | Authors: | Hart, P.J, Deep, S, Taylor, A.B, Shu, Z, Hinck, C.S, Hinck, A.P. | | Deposit date: | 2002-01-18 | | Release date: | 2002-02-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of the human TbetaR2 ectodomain--TGF-beta3 complex.
Nat.Struct.Biol., 9, 2002
|
|
2YL6
 
 | | Inhibition of the pneumococcal virulence factor StrH and molecular insights into N-glycan recognition and hydrolysis | | Descriptor: | 1,2-ETHANEDIOL, 2-{2-[2-2-(METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, BETA-N-ACETYLHEXOSAMINIDASE, ... | | Authors: | Pluvinage, B, Higgins, M.A, Abbott, D.W, Robb, C, Dalia, A.B, Deng, L, Weiser, J.N, Parsons, T.B, Fairbanks, A.J, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2011-05-31 | | Release date: | 2011-09-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Inhibition of the Pneumococcal Virulence Factor Strh and Molecular Insights Into N-Glycan Recognition and Hydrolysis.
Structure, 19, 2011
|
|
2YL9
 
 | | INHIBITION OF THE PNEUMOCOCCAL VIRULENCE FACTOR STRH AND MOLECULAR INSIGHTS INTO N-GLYCAN RECOGNITION AND HYDROLYSIS | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BETA-N-ACETYLHEXOSAMINIDASE | | Authors: | Pluvinage, B, Higgins, M.A, Abbott, D.W, Robb, C, Dalia, A.B, Deng, L, Weiser, J.N, Parsons, T.B, Fairbanks, A.J, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2011-06-01 | | Release date: | 2011-09-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Inhibition of the Pneumococcal Virulence Factor Strh and Molecular Insights Into N-Glycan Recognition and Hydrolysis.
Structure, 19, 2011
|
|
2YL8
 
 | | Inhibition of the pneumococcal virulence factor StrH and molecular insights into N-glycan recognition and hydrolysis | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose, BETA-N-ACETYLHEXOSAMINIDASE, ... | | Authors: | Pluvinage, B, Higgins, M.A, Abbott, D.W, Robb, C, Dalia, A.B, Deng, L, Weiser, J.N, Parsons, T.B, Fairbanks, A.J, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2011-05-31 | | Release date: | 2011-09-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Inhibition of the Pneumococcal Virulence Factor Strh and Molecular Insights Into N-Glycan Recognition and Hydrolysis.
Structure, 19, 2011
|
|
2YL5
 
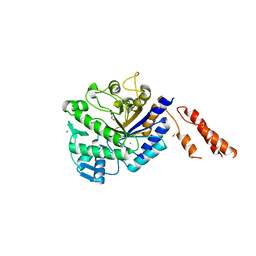 | | Inhibition of the pneumococcal virulence factor StrH and molecular insights into N-glycan recognition and hydrolysis | | Descriptor: | 1,2-ETHANEDIOL, BETA-N-ACETYLHEXOSAMINIDASE, MAGNESIUM ION | | Authors: | Pluvinage, B, Higgins, M.A, Abbott, D.W, Robb, C, Dalia, A.B, Deng, L, Weiser, J.N, Parsons, T.B, Fairbanks, A.J, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2011-05-31 | | Release date: | 2011-09-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Inhibition of the Pneumococcal Virulence Factor Strh and Molecular Insights Into N-Glycan Recognition and Hydrolysis.
Structure, 19, 2011
|
|
2YLL
 
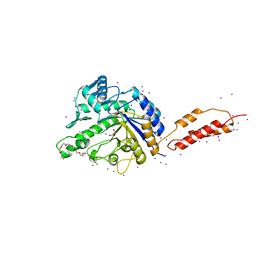 | | Inhibition of the pneumococcal virulence factor StrH and molecular insights into N-glycan recognition and hydrolysis | | Descriptor: | 1,2-ETHANEDIOL, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, BETA-N-ACETYLHEXOSAMINIDASE, ... | | Authors: | Pluvinage, B, Higgins, M.A, Abbott, D.W, Robb, C, Dalia, A.B, Deng, L, Weiser, J.N, Parsons, T.B, Fairbanks, A.J, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2011-06-02 | | Release date: | 2011-09-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Inhibition of the Pneumococcal Virulence Factor Strh and Molecular Insights Into N-Glycan Recognition and Hydrolysis.
Structure, 19, 2011
|
|
2YLA
 
 | | INHIBITION OF THE PNEUMOCOCCAL VIRULENCE FACTOR STRH AND MOLECULAR INSIGHTS INTO N-GLYCAN RECOGNITION AND HYDROLYSIS | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pluvinage, B, Higgins, M.A, Abbott, D.W, Robb, C, Dalia, A.B, Deng, L, Weiser, J.N, Parsons, T.B, Fairbanks, A.J, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2011-06-01 | | Release date: | 2011-09-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Inhibition of the Pneumococcal Virulence Factor Strh and Molecular Insights Into N-Glycan Recognition and Hydrolysis.
Structure, 19, 2011
|
|
9BDL
 
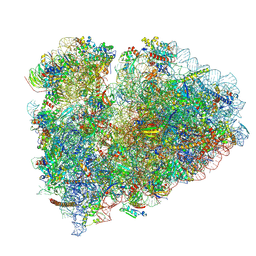 | | 80S ribosome with angiogenin | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S10, ... | | Authors: | Loveland, A.B, Korostelev, A.A. | | Deposit date: | 2024-04-12 | | Release date: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural mechanism of angiogenin activation by the ribosome.
Nature, 630, 2024
|
|
9BNQ
 
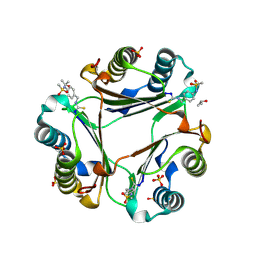 | | N-(4-(isothiocyanatomethyl)phenyl)methanesulfonamide complexed with Macrophage Migration Inhibitory Factor | | Descriptor: | ISOPROPYL ALCOHOL, Macrophage migration inhibitory factor, N-{[4-(methanesulfonamido)phenyl]methyl}methanethioamide, ... | | Authors: | Fellner, M, Rutledge, M.T, Putha, L, Kok, L.K, Gamble, A.B, Wilbanks, S.M, Vernall, A.J, Tyndall, J.D.A. | | Deposit date: | 2024-05-02 | | Release date: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Covalent isothiocyanate inhibitors of macrophage migration inhibitory factor as potential colorectal cancer treatments.
Chemmedchem, 2024
|
|
9BUJ
 
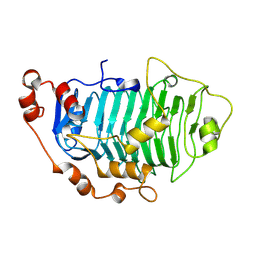 | | Structure of PfPL1 from Pseudoalteromonas fuliginea | | Descriptor: | CALCIUM ION, Pectate lyase | | Authors: | Boraston, A.B, Hobbs, J.K. | | Deposit date: | 2024-05-17 | | Release date: | 2024-07-10 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | The structure of a pectin-active family 1 polysaccharide lyase from the marine bacterium Pseudoalteromonas fuliginea.
Acta Crystallogr.,Sect.F, 80, 2024
|
|
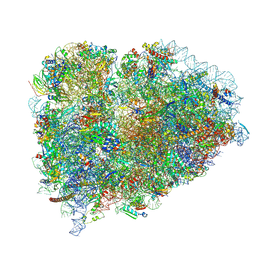
9BDP
 
 | |
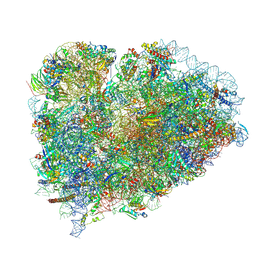
9BDN
 
 | |
9FJK
 
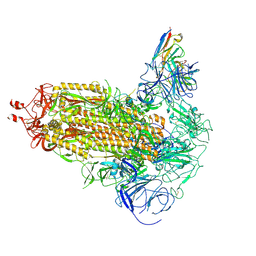 | | Omicron BA.1 Spike protein with neutralizing NTD specific mAb K501SP6 | | Descriptor: | K501SP6 Fv Heavy Chain, K501SP6 Fv Light Chain, Spike glycoprotein,Fibritin | | Authors: | Bjoernsson, K.H, Walker, M.R, Raghavan, S.S.R, Ward, A.B, Barfod, L.K. | | Deposit date: | 2024-05-31 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Omicron BA.1 Spike protein with neutralizing NTD specific mAb K501SP6
To Be Published
|
|
9BNR
 
 | | 4-(2-isothiocyanatoethyl)benzenesulfonamide complexed with Macrophage Migration Inhibitory Factor | | Descriptor: | Macrophage migration inhibitory factor, N-[2-(4-sulfamoylphenyl)ethyl]methanethioamide, SULFATE ION | | Authors: | Fellner, M, Rutledge, M.T, Putha, L, Kok, L.K, Gamble, A.B, Wilbanks, S.M, Vernall, A.J, Tyndall, J.D.A. | | Deposit date: | 2024-05-02 | | Release date: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Covalent isothiocyanate inhibitors of macrophage migration inhibitory factor as potential colorectal cancer treatments.
Chemmedchem, 2024
|
|
