7N5F
 
 | | Structure of Mechanosensitive Ion Channel Flycatcher1 Protomer in 'Down' conformation in GDN | | Descriptor: | Mechanosensitive ion channel Flycatcher1, PALMITIC ACID | | Authors: | Jojoa-Cruz, S, Saotome, K, Lee, W.H, Patapoutian, A, Ward, A.B. | | Deposit date: | 2021-06-05 | | Release date: | 2022-02-16 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural insights into the Venus flytrap mechanosensitive ion channel Flycatcher1.
Nat Commun, 13, 2022
|
|
7N5D
 
 | | Composite Structure of Mechanosensitive Ion Channel Flycatcher1 in GDN | | Descriptor: | Mechanosensitive ion channel Flycatcher1, PALMITIC ACID | | Authors: | Jojoa-Cruz, S, Saotome, K, Lee, W.H, Patapoutian, A, Ward, A.B. | | Deposit date: | 2021-06-05 | | Release date: | 2022-02-16 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural insights into the Venus flytrap mechanosensitive ion channel Flycatcher1.
Nat Commun, 13, 2022
|
|
7N5G
 
 | | Structure of Mechanosensitive Ion Channel Flycatcher1 Protomer in 'Up' conformation in GDN | | Descriptor: | Mechanosensitive ion channel Flycatcher1, PALMITIC ACID | | Authors: | Jojoa-Cruz, S, Saotome, K, Lee, W.H, Patapoutian, A, Ward, A.B. | | Deposit date: | 2021-06-05 | | Release date: | 2022-02-16 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structural insights into the Venus flytrap mechanosensitive ion channel Flycatcher1.
Nat Commun, 13, 2022
|
|
7N5E
 
 | | Structure of Mechanosensitive Ion Channel Flycatcher1 in GDN | | Descriptor: | Mechanosensitive ion channel Flycatcher1, PALMITIC ACID | | Authors: | Jojoa-Cruz, S, Saotome, K, Lee, W.H, Patapoutian, A, Ward, A.B. | | Deposit date: | 2021-06-05 | | Release date: | 2022-02-16 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural insights into the Venus flytrap mechanosensitive ion channel Flycatcher1.
Nat Commun, 13, 2022
|
|
7LV0
 
 | | Pre-translocation rotated ribosome +1-frameshifting(CCC-A) complex (Structure Irot-FS) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Demo, G, Loveland, A.B, Svidritskiy, E, Gamper, H.B, Hou, Y.M, Korostelev, A.A. | | Deposit date: | 2021-02-23 | | Release date: | 2021-07-28 | | Last modified: | 2021-08-25 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for +1 ribosomal frameshifting during EF-G-catalyzed translocation.
Nat Commun, 12, 2021
|
|
7LG6
 
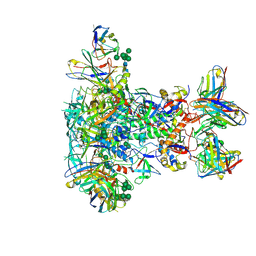 | | BG505 SOSIP.v5.2 in complex with VRC40.01 and RM19R Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | Authors: | Cottrell, C.A, Torres, J.L, Wu, N.R, Ward, A.B. | | Deposit date: | 2021-01-19 | | Release date: | 2021-09-15 | | Last modified: | 2021-11-17 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Structural basis of glycan276-dependent recognition by HIV-1 broadly neutralizing antibodies.
Cell Rep, 37, 2021
|
|
7JVI
 
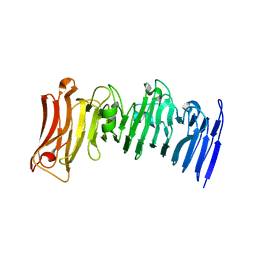 | | Crystal structure of a beta-helix domain retrieved from capybara gut metagenome | | Descriptor: | Beta-helix domain, CALCIUM ION | | Authors: | Martins, M.P, Genoroso, W.C, Domingues, M.N, Persinoti, G.F, Morais, M.A.B, Murakami, M.T. | | Deposit date: | 2020-08-21 | | Release date: | 2021-09-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Gut microbiome of the largest living rodent harbors unprecedented enzymatic systems to degrade plant polysaccharides.
Nat Commun, 13, 2022
|
|
7JVH
 
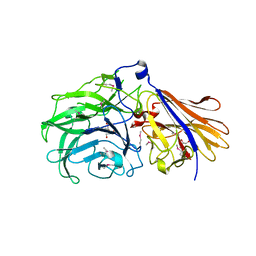 | | Crystal structure of a GH43_12 retrieved from capybara gut metagenome | | Descriptor: | GLYCEROL, Glycoside Hydrolase Family 43_12 | | Authors: | Cabral, L, Domingues, M.N, Martins, M.P, Persinoti, G.F, Morais, M.A.B, Murakami, M.T. | | Deposit date: | 2020-08-21 | | Release date: | 2021-09-08 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Crystal structure of a GH43_12 retrieved from capybara gut metagenome
To Be Published
|
|
7MGQ
 
 | | AICAR transformylase/IMP cyclohydrolase (ATIC) is essential for de novo purine biosynthesis and infection by Cryptococcus neoformans | | Descriptor: | 5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase, MAGNESIUM ION | | Authors: | Wizrah, M.S, Chua, S.M.H, Luo, Z, Manik, M.K, Pan, M, Whyte, J.M, Robertson, A.B, Kappler, U, Kobe, B, Fraser, J.A. | | Deposit date: | 2021-04-13 | | Release date: | 2022-04-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | AICAR transformylase/IMP cyclohydrolase (ATIC) is essential for de novo purine biosynthesis and infection by Cryptococcus neoformans.
J.Biol.Chem., 298, 2022
|
|
7MLH
 
 | | Crystal structure of human IgE (2F10) in complex with Der p 2.0103 | | Descriptor: | CHLORIDE ION, Der p 2 variant 3, IgE Heavy chain, ... | | Authors: | Khatri, K, Kapingidza, A.B, Richardson, C.M, Vailes, L.D, Wunschmann, S, Dolamore, C, Chapman, M.D, Smith, S.A, Pomes, A, Chruszcz, M. | | Deposit date: | 2021-04-28 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Human IgE monoclonal antibody recognition of mite allergen Der p 2 defines structural basis of an epitope for IgE cross-linking and anaphylaxis in vivo.
Pnas Nexus, 1, 2022
|
|
7O5H
 
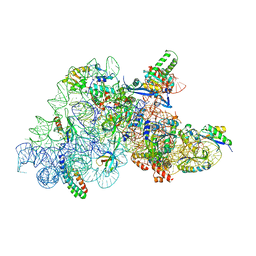 | | Ribosomal methyltransferase KsgA bound to small ribosomal subunit | | Descriptor: | 16S rRNA, 30S ribosomal protein S11, 30S ribosomal protein S12, ... | | Authors: | Stephan, N.C, Ries, A.B, Boehringer, D, Ban, N. | | Deposit date: | 2021-04-08 | | Release date: | 2021-06-16 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of successive adenosine modifications by the conserved ribosomal methyltransferase KsgA.
Nucleic Acids Res., 49, 2021
|
|
1OQH
 
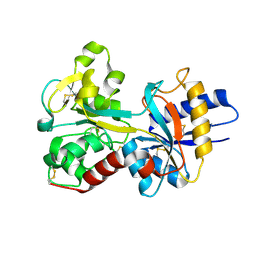 | | Crystal Structure of the R124A mutant of the N-lobe human transferrin | | Descriptor: | CARBONATE ION, FE (III) ION, POTASSIUM ION, ... | | Authors: | Baker, H.M, He, Q.-Y, Brigg, S.K, Mason, A.B, N Baker, E. | | Deposit date: | 2003-03-10 | | Release date: | 2003-03-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and functional consequences of binding site mutations in transferrin: crystal structures of the Asp63Glu and Arg124Ala mutants of the N-lobe of human transferrin
Biochemistry, 42, 2003
|
|
2OSF
 
 | | Inhibition of Carbonic Anhydrase II by Thioxolone: A Mechanistic and Structural Study | | Descriptor: | 4-MERCAPTOBENZENE-1,3-DIOL, Carbonic anhydrase 2, S-(2,4-dihydroxyphenyl) hydrogen thiocarbonate, ... | | Authors: | Albert, A.B, Caroli, G, Govindasamy, L, Agbandje-McKenna, M, McKenna, R, Tripp, B.C. | | Deposit date: | 2007-02-05 | | Release date: | 2008-05-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Inhibition of carbonic anhydrase II by thioxolone: a mechanistic and structural study.
Biochemistry, 47, 2008
|
|
6SL4
 
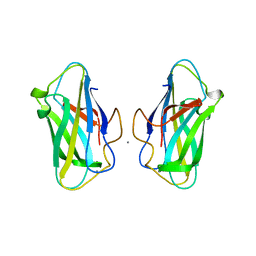 | | The unique CBM-Cthe_0271 of Ruminiclostridium thermocellum | | Descriptor: | CALCIUM ION, Type 3a cellulose-binding domain protein | | Authors: | Milana, M.V, Almog, R, Yaniv, O, Oded, L, Inna, R.G, Felix, F, Edward, A.B, Raphael, L. | | Deposit date: | 2019-08-18 | | Release date: | 2020-03-18 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The unique CBM-Cthe_0271 of Ruminiclostridium thermocellum
To Be Published
|
|
1P8D
 
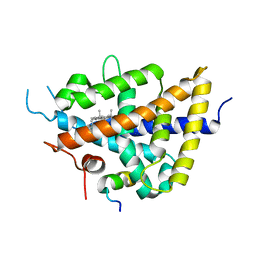 | | X-Ray Crystal Structure of LXR Ligand Binding Domain with 24(S),25-epoxycholesterol | | Descriptor: | 17-[3-(3,3-DIMETHYL-OXIRANYL)-1-METHYL-PROPYL]-10,13-DIMETHYL-2,3,4,7,8,9,10,11,12,13,14,15,16,17-TETRADECAHYDRO-1H-CYC LOPENTA[A]PHENANTHREN-3-OL, Oxysterols receptor LXR-beta, nuclear receptor coactivator 1 isoform 3 | | Authors: | Williams, S, Bledsoe, R.K, Collins, J.L, Boggs, S, Lambert, M.H, Miller, A.B, Moore, J, McKee, D.D, Moore, L, Nichols, J, Parks, D, Watson, M, Wisely, B, Willson, T.M. | | Deposit date: | 2003-05-06 | | Release date: | 2003-07-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-ray crystal structure of the liver X receptor beta ligand binding domain: regulation by
a histidine-tryptophan switch.
J.Biol.Chem., 278, 2003
|
|
2QWQ
 
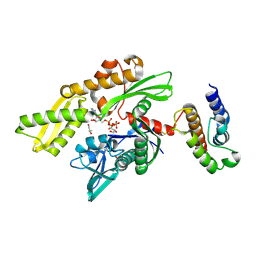 | | Crystal structure of disulfide-bond-crosslinked complex of bovine hsc70 (1-394aa)R171C and bovine Auxilin (810-910aa)D876C in the AMPPNP hydrolyzed form | | Descriptor: | ACETIC ACID, ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, ... | | Authors: | Jiang, J, Maes, E.G, Wang, L, Taylor, A.B, Hinck, A.P, Lafer, E.M, Sousa, R. | | Deposit date: | 2007-08-10 | | Release date: | 2007-12-18 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structural basis of J cochaperone binding and regulation of Hsp70.
Mol.Cell, 28, 2007
|
|
5ZBU
 
 | |
6U54
 
 | | Anti-Zaire ebolavirus Nucleoprotein Single Domain Antibody Zaire C (ZC) Complexed with Zaire ebolavirus Nucleoprotein C-terminal Domain 634-739 | | Descriptor: | Anti-Zaire ebolavirus Nucleoprotein Single Domain Antibody Zaire C (ZC), Nucleoprotein | | Authors: | Taylor, A.B, Sherwood, L.J, Hart, P.J, Hayhurst, A. | | Deposit date: | 2019-08-26 | | Release date: | 2019-11-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Paratope Duality and Gullying are Among the Atypical Recognition Mechanisms Used by a Trio of Nanobodies to Differentiate Ebolavirus Nucleoproteins.
J.Mol.Biol., 431, 2019
|
|
1QZQ
 
 | | human Tyrosyl DNA phosphodiesterase | | Descriptor: | tyrosyl-DNA phosphodiesterase 1 | | Authors: | Raymond, A.C, Rideout, M.C, Staker, B, Hjerrild, K, Burgin Jr, A.B. | | Deposit date: | 2003-09-17 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Analysis of Human Tyrosyl-DNA Phosphodiesterase I Catalytic Residues.
J.Mol.Biol., 338, 2004
|
|
1QVS
 
 | | Crystal Structure of Haemophilus influenzae H9A mutant Holo Ferric ion-Binding Protein A | | Descriptor: | FE (III) ION, Iron-utilization periplasmic protein, PHOSPHATE ION | | Authors: | Shouldice, S.R, Skene, R.J, Dougan, D.R, McRee, D.E, Tari, L.W, Schryvers, A.B. | | Deposit date: | 2003-08-28 | | Release date: | 2003-11-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Presence of ferric hydroxide clusters in mutants of Haemophilus influenzae ferric ion-binding protein A
Biochemistry, 42, 2003
|
|
1QW0
 
 | | Crystal Structure of Haemophilus influenzae N175L mutant Holo Ferric ion-Binding Protein A | | Descriptor: | FE (III) ION, Iron-utilization periplasmic protein, PHOSPHATE ION | | Authors: | Shouldice, S.R, Skene, R.J, Dougan, D.R, McRee, D.E, Tari, L.W, Schryvers, A.B. | | Deposit date: | 2003-08-29 | | Release date: | 2003-11-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Presence of ferric hydroxide clusters in mutants of Haemophilus influenzae ferric ion-binding protein A
Biochemistry, 42, 2003
|
|
6U02
 
 | | CryoEM-derived model of NA-63 Fab in complex with N9 Shanghai2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab-63 Heavy Chain, Fab-63 Light Chain, ... | | Authors: | Ward, A.B, Turner, H.L, Zhu, X. | | Deposit date: | 2019-08-13 | | Release date: | 2019-12-04 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Structural Basis of Protection against H7N9 Influenza Virus by Human Anti-N9 Neuraminidase Antibodies.
Cell Host Microbe, 26, 2019
|
|
6UGQ
 
 | | Human Carbonic Anhydrase IX-mimic complexed with SB4-206 | | Descriptor: | 5-chloro-1lambda~6~,2,4-benzothiadiazine-1,1,3(2H,4H)-trione, Carbonic anhydrase IX-mimic, ZINC ION | | Authors: | Murray, A.B, Lomelino, C.L, McKenna, R. | | Deposit date: | 2019-09-26 | | Release date: | 2019-12-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.305 Å) | | Cite: | "A Sweet Combination": Developing Saccharin and Acesulfame K Structures for Selectively Targeting the Tumor-Associated Carbonic Anhydrases IX and XII.
J.Med.Chem., 63, 2020
|
|
1OI6
 
 | | Structure determination of the TMP-complex of EvaD | | Descriptor: | GLYCEROL, PCZA361.16, THYMIDINE-5'-PHOSPHATE | | Authors: | Merkel, A.B, Naismith, J.H. | | Deposit date: | 2003-06-09 | | Release date: | 2004-06-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Position of a Key Tyrosine in Dtdp-4-Keto-6-Deoxy-D-Glucose-5-Epimerase (Evad) Alters the Substrate Profile for This Rmlc-Like Enzyme
J.Biol.Chem., 279, 2004
|
|
6U4T
 
 | | Carbonic anhydrase 9 in complex with SB4197 | | Descriptor: | 4-methyl-1lambda~6~,2,4-benzothiadiazine-1,1,3(2H,4H)-trione, Carbonic anhydrase 2, ZINC ION | | Authors: | Murray, A.B, Lomelino, C.L, McKenna, R. | | Deposit date: | 2019-08-26 | | Release date: | 2020-01-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.356 Å) | | Cite: | "A Sweet Combination": Developing Saccharin and Acesulfame K Structures for Selectively Targeting the Tumor-Associated Carbonic Anhydrases IX and XII.
J.Med.Chem., 63, 2020
|
|
