4OOI
 
 | | Reduced HlyU from Vibrio cholerae N16961 | | Descriptor: | Transcriptional activator HlyU | | Authors: | Mukherjee, D, Datta, A.B, Chakrabarti, P. | | Deposit date: | 2014-02-03 | | Release date: | 2014-12-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of HlyU, the hemolysin gene transcription activator, from Vibrio cholerae N16961 and functional implications
Biochim.Biophys.Acta, 1844, 2014
|
|
6XBR
 
 | | Streptomyces coelicolor methylmalonyl-CoA epimerase (E43L) in complex with 2-nitronate-propionyl-CoA | | Descriptor: | AMMONIUM ION, CHLORIDE ION, COBALT (II) ION, ... | | Authors: | Stunkard, L.M, Boram, T.J, Benjamin, A.B, Bower, J.B, Lohman, J.R. | | Deposit date: | 2020-06-07 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Streptomyces coelicolor methylmalonyl-CoA epimerase (E43L) in complex with 2-nitronate-propionyl-CoA
To Be Published
|
|
4OYQ
 
 | | (6-isothiocyanatohexyl)benzene inhibitor complexed with Macrophage Migration Inhibitory Factor | | Descriptor: | 6-isothiocyanatohexylbenzene, GLYCEROL, Macrophage migration inhibitory factor, ... | | Authors: | Spencer, E.S, Dale, E.J, Gommans, A.L, Vo, C.T, Rutledge, M.T, Nakatani, Y, Gamble, A.B, Smith, R.A.J, Wilbanks, S.M, Hampton, M.B, Tyndall, J.D.A. | | Deposit date: | 2014-02-12 | | Release date: | 2014-03-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | To be published
To be published
|
|
6XBV
 
 | | Streptomyces coelicolor methylmalonyl-CoA epimerase (S115T) in complex with 2-nitronate-propionyl-CoA | | Descriptor: | CHLORIDE ION, COBALT (II) ION, Methylmalonyl-CoA epimerase, ... | | Authors: | Stunkard, L.M, Boram, T.J, Benjamin, A.B, Bower, J.B, Lohman, J.R. | | Deposit date: | 2020-06-07 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Streptomyces coelicolor methylmalonyl-CoA epimerase (S115T) in complex with 2-nitronate-propionyl-CoA
To Be Published
|
|
5LGF
 
 | |
7JFS
 
 | |
4OSF
 
 | | 4-(2-isothiocyanatoethyl)phenol inhibitor complexed with Macrophage Migration Inhibitory Factor | | Descriptor: | CHLORIDE ION, ISOPROPYL ALCOHOL, Macrophage migration inhibitory factor, ... | | Authors: | Spencer, E.S, Dale, E.J, Gommans, A.L, Vo, C.T, Rutledge, M.T, Nakatani, Y, Gamble, A.B, Smith, R.A.J, Wilbanks, S.M, Hampton, M.B, Tyndall, J.D.A. | | Deposit date: | 2014-02-12 | | Release date: | 2014-03-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Multiple binding modes of isothiocyanates that inhibit macrophage migration inhibitory factor
Eur.J.Med.Chem., 93, 2015
|
|
6OG7
 
 | | 70S termination complex with RF2 bound to the UGA codon. Non-rotated ribosome with RF2 bound (Structure II) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Svidritskiy, E, Demo, G, Loveland, A.B, Xu, C, Korostelev, A.A. | | Deposit date: | 2019-04-01 | | Release date: | 2019-09-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Extensive ribosome and RF2 rearrangements during translation termination.
Elife, 8, 2019
|
|
6XX1
 
 | | The unique CBM3-Clocl_1192 of Hungateiclostridium clariflavum | | Descriptor: | Cellulose binding domain-containing protein | | Authors: | Milana, M.V, Almog, R, Yaniv, O, Oded, L, Inna, R.G, Felix, F, Edward, A.B, Raphael, L. | | Deposit date: | 2020-01-26 | | Release date: | 2020-05-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | The unique CBM3-Cthe_0271 from Ruminoclostridium thermocellum and CBM3-Clocl_1192 from Hungateiclostridium clariflavum
To Be Published
|
|
6XT1
 
 | |
4PEI
 
 | | Dbr1 in complex with synthetic branched RNA analog | | Descriptor: | GLYCEROL, NICKEL (II) ION, RNA (5'-R(*(G46)P*U)-3'), ... | | Authors: | Montemayor, E.J, Katolik, A, Clark, N.E, Taylor, A.B, Schuermann, J.P, Combs, D.J, Johnsson, R, Holloway, S.P, Stevens, S.W, Damha, M.J, Hart, P.J. | | Deposit date: | 2014-04-23 | | Release date: | 2014-08-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis of lariat RNA recognition by the intron debranching enzyme Dbr1.
Nucleic Acids Res., 42, 2014
|
|
4PEG
 
 | | Dbr1 in complex with guanosine-5'-monophosphate | | Descriptor: | GLYCEROL, GUANOSINE-5'-MONOPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Montemayor, E.J, Katolik, A, Clark, N.E, Taylor, A.B, Schuermann, J.P, Combs, D.J, Johnsson, R, Holloway, S.P, Stevens, S.W, Damha, M.J, Hart, P.J. | | Deposit date: | 2014-04-23 | | Release date: | 2014-08-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of lariat RNA recognition by the intron debranching enzyme Dbr1.
Nucleic Acids Res., 42, 2014
|
|
6OHY
 
 | | Chimpanzee SIV Env trimeric ectodomain. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pallesen, J, Andrabi, R, de Val, N, Burton, D.R, Ward, A.B. | | Deposit date: | 2019-04-08 | | Release date: | 2019-06-05 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | The Chimpanzee SIV Envelope Trimer: Structure and Deployment as an HIV Vaccine Template.
Cell Rep, 27, 2019
|
|
4PHC
 
 | | Crystal Structure of a human cytosolic histidyl-tRNA synthetase, histidine-bound | | Descriptor: | GLYCEROL, HISTIDINE, Histidine--tRNA ligase, ... | | Authors: | Koh, C.Y, Wetzel, A.B, de van der Schueren, W.J, Hol, W.G.J. | | Deposit date: | 2014-05-06 | | Release date: | 2014-08-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.844 Å) | | Cite: | Comparison of histidine recognition in human and trypanosomatid histidyl-tRNA synthetases.
Biochimie, 106, 2014
|
|
6X7Q
 
 | | Chloramphenicol acetyltransferase type III in complex with chloramphenicol and acetyl-oxa(dethia)-CoA | | Descriptor: | CHLORAMPHENICOL, Chloramphenicol acetyltransferase 3, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Benjamin, A.B, Stunkard, L.M, Ling, J, Nice, J.N, Lohman, J.R. | | Deposit date: | 2020-05-30 | | Release date: | 2021-06-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structures of chloramphenicol acetyltransferase III and Escherichia coli beta-keto-acylsynthase III co-crystallized with partially hydrolysed acetyl-oxa(de-thia)CoA
Acta Crystallogr.,Sect.F, 2023
|
|
6X7R
 
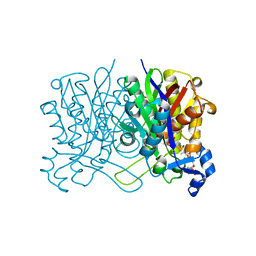 | | E. coli beta-ketoacyl-[acyl carrier protein] synthase III (FabH) in complex with oxa(dethia)-coenzyme A | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase 3, DIMETHYL SULFOXIDE, MAGNESIUM ION, ... | | Authors: | Benjamin, A.B, Stunkard, L.M, Ling, J, Nice, J.N, Lohman, J.R. | | Deposit date: | 2020-05-30 | | Release date: | 2021-06-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structures of chloramphenicol acetyltransferase III and Escherichia coli beta-keto-acylsynthase III co-crystallized with partially hydrolysed acetyl-oxa(de-thia)CoA
Acta Crystallogr.,Sect.F, 2023
|
|
5LME
 
 | | Specific-DNA binding activity of the cross-brace zinc finger motif of the piggyBac transposase | | Descriptor: | ZINC ION, piggyBac transposase | | Authors: | Morellet, N, Taylor, J.A, Wieninger, S, Moriau, S, Li, X, Lescop, E, Mathy, N, Bischerour, J, Betermier, M, Bardiaux, B, Nilges, M, Craig, N.L, Hickman, A.B, Dyda, F, Guittet, E. | | Deposit date: | 2016-07-30 | | Release date: | 2017-12-20 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Sequence-specific DNA binding activity of the cross-brace zinc finger motif of the piggyBac transposase.
Nucleic Acids Res., 46, 2018
|
|
6OFX
 
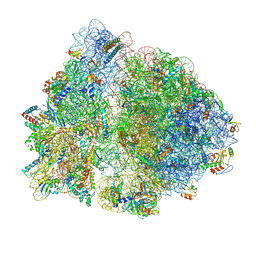 | | Non-rotated ribosome (Structure I) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Svidritskiy, E, Demo, G, Loveland, A.B, Xu, C, Korostelev, A.A. | | Deposit date: | 2019-04-01 | | Release date: | 2019-09-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Extensive ribosome and RF2 rearrangements during translation termination.
Elife, 8, 2019
|
|
6ORH
 
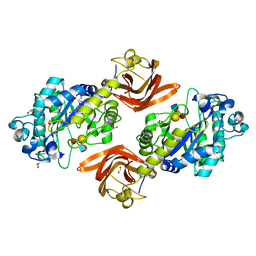 | | Crystal structure of SpGH29 | | Descriptor: | 1,2-ETHANEDIOL, Glycoside hydrolase, alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-alpha-D-glucopyranose | | Authors: | Pluvinage, B, Boraston, A.B. | | Deposit date: | 2019-04-30 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Two complementary alpha-fucosidases fromStreptococcus pneumoniaepromote complete degradation of host-derived carbohydrate antigens.
J.Biol.Chem., 294, 2019
|
|
4PEF
 
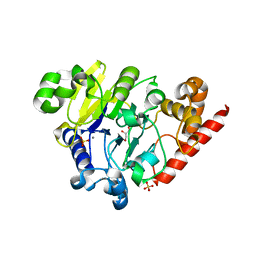 | | Dbr1 in complex with sulfate | | Descriptor: | GLYCEROL, MANGANESE (II) ION, RNA lariat debranching enzyme, ... | | Authors: | Montemayor, E.J, Katolik, A, Clark, N.E, Taylor, A.B, Schuermann, J.P, Combs, D.J, Johnsson, R, Holloway, S.P, Stevens, S.W, Damha, M.J, Hart, P.J. | | Deposit date: | 2014-04-23 | | Release date: | 2014-08-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural basis of lariat RNA recognition by the intron debranching enzyme Dbr1.
Nucleic Acids Res., 42, 2014
|
|
6OY4
 
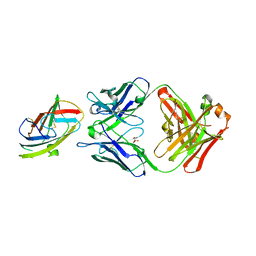 | | Crystal structure of complex between recombinant Der p 2.0103 and Fab fragment of 7A1 | | Descriptor: | Der p 2 variant 3, Fab fragment of IgG, HEAVY CHAIN, ... | | Authors: | Kapingidza, A.B, Offermann, L.R, Glesner, J, Wunschmann, S, Vailes, L.D, Chapman, M.D.C, Pomes, A, Chruszcz, M. | | Deposit date: | 2019-05-14 | | Release date: | 2019-08-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | A Human IgE Antibody Binding Site on Der p 2 for the Design of a Recombinant Allergen for Immunotherapy.
J Immunol., 203, 2019
|
|
7JNB
 
 | |
7JS4
 
 | |
6X96
 
 | |
6X98
 
 | |
