4RNU
 
 | | G303 Circular Permutation of Old Yellow Enzyme | | 分子名称: | FLAVIN MONONUCLEOTIDE, NADPH dehydrogenase 1, PHOSPHATE ION | | 著者 | Horton, J.R, Daugherty, A.B, Cheng, X, Lutz, S. | | 登録日 | 2014-10-26 | | 公開日 | 2015-01-14 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.677 Å) | | 主引用文献 | STRUCTURAL AND FUNCTIONAL CONSEQUENCES OF CIRCULAR PERMUTATION ON THE ACTIVE SITE OF OLD YELLOW ENZYME.
ACS Catal, 5, 2015
|
|
4RP6
 
 | |
3GS2
 
 | | Ring1B C-terminal domain/Cbx7 Cbox Complex | | 分子名称: | Chromobox protein homolog 7, E3 ubiquitin-protein ligase RING2, SULFATE ION, ... | | 著者 | Wang, R, Taylor, A.B, Kim, C.A. | | 登録日 | 2009-03-26 | | 公開日 | 2010-08-25 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.699 Å) | | 主引用文献 | Polycomb Group Targeting through Different Binding Partners of RING1B C-Terminal Domain.
Structure, 18, 2010
|
|
3H2P
 
 | | Human SOD1 D124V Variant | | 分子名称: | ACETYL GROUP, MALONATE ION, Superoxide dismutase [Cu-Zn], ... | | 著者 | Seetharaman, S.V, Winkler, D.D, Taylor, A.B, Cao, X, Whitson, L.J, Doucette, P.A, Valentine, J.S, Carroll, M.C, Culotta, V.C, Hart, P.J. | | 登録日 | 2009-04-14 | | 公開日 | 2010-05-05 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Structures of Pathogenic SOD1 Mutants H80R and D124V: Disrupted Zinc-binding
and Compromised Post-translational Modification by the Copper Chaperone CCS
To be Published
|
|
5KPX
 
 | | Structure of RelA bound to ribosome in presence of A/R tRNA (Structure IV) | | 分子名称: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Loveland, A.B, Bah, E, Madireddy, R, Zhang, Y, Brilot, A.F, Grigorieff, N, Korostelev, A.A. | | 登録日 | 2016-07-05 | | 公開日 | 2016-09-28 | | 最終更新日 | 2019-11-20 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Ribosome•RelA structures reveal the mechanism of stringent response activation.
Elife, 5, 2016
|
|
4RNV
 
 | | G303 Circular Permutation of Old Yellow Enzyme with the Inhibitor p-Hydroxybenzaldehyde | | 分子名称: | FLAVIN MONONUCLEOTIDE, NADPH dehydrogenase 1, P-HYDROXYBENZALDEHYDE | | 著者 | Horton, J.R, Daugherty, A.B, Cheng, X, Lutz, S. | | 登録日 | 2014-10-26 | | 公開日 | 2015-01-14 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.473 Å) | | 主引用文献 | STRUCTURAL AND FUNCTIONAL CONSEQUENCES OF CIRCULAR PERMUTATION ON THE ACTIVE SITE OF OLD YELLOW ENZYME.
ACS Catal, 5, 2015
|
|
3B7A
 
 | |
3B88
 
 | |
3GTV
 
 | |
3BBH
 
 | | M. jannaschii Nep1 complexed with Sinefungin | | 分子名称: | GLYCEROL, Ribosome biogenesis protein NEP1-like, SINEFUNGIN | | 著者 | Taylor, A.B, Meyer, B, Leal, B.Z, Kotter, P, Hart, P.J, Entian, K.-D, Wohnert, J. | | 登録日 | 2007-11-09 | | 公開日 | 2008-02-05 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | The crystal structure of Nep1 reveals an extended SPOUT-class methyltransferase fold and a pre-organized SAM-binding site.
Nucleic Acids Res., 36, 2008
|
|
3BBE
 
 | | M. jannaschii Nep1 | | 分子名称: | GLYCEROL, Ribosome biogenesis protein NEP1-like | | 著者 | Taylor, A.B, Meyer, B, Leal, B.Z, Kotter, P, Hart, P.J, Entian, K.-D, Wohnert, J. | | 登録日 | 2007-11-09 | | 公開日 | 2008-02-05 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | The crystal structure of Nep1 reveals an extended SPOUT-class methyltransferase fold and a pre-organized SAM-binding site.
Nucleic Acids Res., 36, 2008
|
|
7QWV
 
 | | Crystal structure of the REC114-TOPOVIBL complex. | | 分子名称: | Meiotic recombination protein REC114, Type 2 DNA topoisomerase 6 subunit B-like | | 著者 | Juarez-Martinez, A.B, Robert, T, de Massy, B, Kadlec, J. | | 登録日 | 2022-01-25 | | 公開日 | 2023-02-01 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.26 Å) | | 主引用文献 | TOPOVIBL-REC114 interaction regulates meiotic DNA double-strand breaks.
Nat Commun, 13, 2022
|
|
4TXW
 
 | | Crystal structure of CBM32-4 from the Clostridium perfringens NagH | | 分子名称: | 1,2-ETHANEDIOL, CALCIUM ION, Hyaluronoglucosaminidase | | 著者 | Grondin, J.M, Ficko-Blean, E, Boraston, A.B, Smith, S.P. | | 登録日 | 2014-07-07 | | 公開日 | 2015-12-09 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Solution Structure and Dynamics of Full-length GH84A, a multimodular B-N-acetylglucosaminidase from Clostridium perfringens
To Be Published
|
|
4U4S
 
 | | Crystal structure of the GluA2 ligand-binding domain (S1S2J-L483Y-N754S) in complex with glutamate and BPAM25 at 1.90 A resolution. | | 分子名称: | 4-ethyl-3,4-dihydro-2H-pyrido[4,3-e][1,2,4]thiadiazine 1,1-dioxide, ACETATE ION, CHLORIDE ION, ... | | 著者 | Noerholm, A.B, Deva, T, Frydenvang, K, Kastrup, J.S. | | 登録日 | 2014-07-24 | | 公開日 | 2014-11-19 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Positive Allosteric Modulators of 2-Amino-3-(3-hydroxy-5-methylisoxazol-4-yl)propionic Acid Receptors Belonging to 4-Cyclopropyl-3,4-dihydro-2H-1,2,4-pyridothiadiazine Dioxides and Diversely Chloro-Substituted 4-Cyclopropyl-3,4-dihydro-2H-1,2,4-benzothiadiazine 1,1-Dioxides.
J.Med.Chem., 57, 2014
|
|
4U4X
 
 | | Crystal structure of the GluA2 ligand-binding domain (S1S2J-L483Y-N754S) in complex with glutamate and BPAM37 at 1.56 A resolution. | | 分子名称: | 4-ethyl-3,4-dihydro-2H-pyrido[3,2-e][1,2,4]thiadiazine 1,1-dioxide, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Noerholm, A.B, Frydenvang, K, Kastrup, J.S. | | 登録日 | 2014-07-24 | | 公開日 | 2014-11-19 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.56 Å) | | 主引用文献 | Positive Allosteric Modulators of 2-Amino-3-(3-hydroxy-5-methylisoxazol-4-yl)propionic Acid Receptors Belonging to 4-Cyclopropyl-3,4-dihydro-2H-1,2,4-pyridothiadiazine Dioxides and Diversely Chloro-Substituted 4-Cyclopropyl-3,4-dihydro-2H-1,2,4-benzothiadiazine 1,1-Dioxides.
J.Med.Chem., 57, 2014
|
|
3BLV
 
 | | Yeast Isocitrate Dehydrogenase with Citrate Bound in the Regulatory Subunits | | 分子名称: | CITRATE ANION, Isocitrate dehydrogenase [NAD] subunit 1, Isocitrate dehydrogenase [NAD] subunit 2 | | 著者 | Taylor, A.B, Hu, G, Hart, P.J, McAlister-Henn, L. | | 登録日 | 2007-12-11 | | 公開日 | 2008-02-05 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Allosteric Motions in Structures of Yeast NAD+-specific Isocitrate Dehydrogenase.
J.Biol.Chem., 283, 2008
|
|
3HOG
 
 | |
7RSO
 
 | | AMC016 SOSIP.v4.2 in complex with PGV04 Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, AMC016 gp120, ... | | 著者 | Bader, D.L.V, Cottrell, C.A, Ward, A.B. | | 登録日 | 2021-08-11 | | 公開日 | 2021-09-15 | | 最終更新日 | 2022-01-26 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | The Glycan Hole Area of HIV-1 Envelope Trimers Contributes Prominently to the Induction of Autologous Neutralization.
J.Virol., 96, 2022
|
|
7RSN
 
 | | AMC018 SOSIP.v4.2 in complex with PGV04 Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, AMC018 gp120, ... | | 著者 | Cottrell, C.A, Ward, A.B. | | 登録日 | 2021-08-11 | | 公開日 | 2021-09-15 | | 最終更新日 | 2022-01-26 | | 実験手法 | ELECTRON MICROSCOPY (3.49 Å) | | 主引用文献 | The Glycan Hole Area of HIV-1 Envelope Trimers Contributes Prominently to the Induction of Autologous Neutralization.
J.Virol., 96, 2022
|
|
4UAP
 
 | | X-ray structure of GH31 CBM32-2 bound to GalNAc | | 分子名称: | 2-acetamido-2-deoxy-beta-D-galactopyranose, CALCIUM ION, GLYCEROL, ... | | 著者 | Grondin, J.M, Abe, K, Boraston, A.B, Smith, S.P. | | 登録日 | 2014-08-11 | | 公開日 | 2015-10-07 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Diverse modes of galacto-specific carbohydrate recognition by a family 31 glycoside hydrolase from Clostridium perfringens.
PLoS ONE, 12, 2017
|
|
4UCU
 
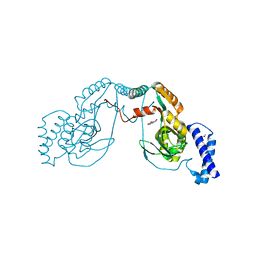 | | Fragment bound to H.influenza NAD dependent DNA ligase | | 分子名称: | 1-(2,4-dimethylbenzyl)-6-oxo-1,6-dihydropyridine-3-carboxamide, 8-hydroxyquinoline-2-carboxylic acid, DNA LIGASE | | 著者 | Hale, M, Brassington, C, Carcanague, D, Embrey, K, Eyermann, C.J, Giacobbe, R.A, Gingipali, L, Gowravaram, M, Harang, J, Howard, T, Ioannidis, G, Jahic, H, Kutschke, A, Laganas, V.A, Loch, J, Miller, M.D, Murphy-Benenato, K.E, Oguto, H, Otterbein, L, Patel, S.J, Shapiro, A.B, Boriack-Sjodin, P.A. | | 登録日 | 2014-12-04 | | 公開日 | 2015-10-14 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | From Fragments to Leads: Novel Bacterial Nad+-Dependent DNA Ligase Inhibitors
Tetrahedron Lett., 56, 2015
|
|
3HOE
 
 | |
3HOL
 
 | |
3PZZ
 
 | |
4UCT
 
 | | Fragment bound to H.influenza NAD dependent DNA ligase | | 分子名称: | 1-(2,4-dimethylbenzyl)-6-oxo-1,6-dihydropyridine-3-carboxamide, 2-amino-6-methyl-5-(propan-2-yloxy)-3H-[1,2,4]triazolo[1,5-a]pyrimidin-8-ium, DNA LIGASE | | 著者 | Hale, M, Brassington, C, Carcanague, D, Embrey, K, Eyermann, C.J, Giacobbe, R.A, Gingipali, L, Gowravaram, M, Harang, J, Howard, T, Ioannidis, G, Jahic, H, Kutschke, A, Laganas, V.A, Loch, J, Miller, M.D, Murphy-Benenato, K.E, Oguto, H, Otterbein, L, Patel, S.J, Shapiro, A.B, Boriack-Sjodin, P.A. | | 登録日 | 2014-12-04 | | 公開日 | 2015-10-14 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | From Fragments to Leads: Novel Bacterial Nad+-Dependent DNA Ligase Inhibitors
Tetrahedron Lett., 56, 2015
|
|
