8VE8
 
 | | Lineage IV Lassa virus glycoprotein (Josiah) in complex with rabbit polyclonal antibody (GP1-A epitope) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Brouwer, P.J.M, Perrett, H.R, Ward, A.B. | | Deposit date: | 2023-12-18 | | Release date: | 2024-09-25 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Defining bottlenecks and opportunities for Lassa virus neutralization by structural profiling of vaccine-induced polyclonal antibody responses.
Cell Rep, 43, 2024
|
|
5KPX
 
 | | Structure of RelA bound to ribosome in presence of A/R tRNA (Structure IV) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Loveland, A.B, Bah, E, Madireddy, R, Zhang, Y, Brilot, A.F, Grigorieff, N, Korostelev, A.A. | | Deposit date: | 2016-07-05 | | Release date: | 2016-09-28 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Ribosome•RelA structures reveal the mechanism of stringent response activation.
Elife, 5, 2016
|
|
6MHG
 
 | |
8VDG
 
 | |
8VDF
 
 | |
8VA7
 
 | | Crystal structure of CapGH3a enzyme retrieved from capybara gut metagenome | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Glycoside hydrolase family 3, ... | | Authors: | Martins, M.P, Vieira, P.S, Morais, M.A.B, Mandelli, F, Chinaglia, M, Lima, E.A, Murakami, M.T. | | Deposit date: | 2023-12-11 | | Release date: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A functionally augmented carbohydrate utilization locus from herbivore gut microbiota fueled by dietary beta-glucans.
NPJ Biofilms Microbiomes, 10, 2024
|
|
8VA4
 
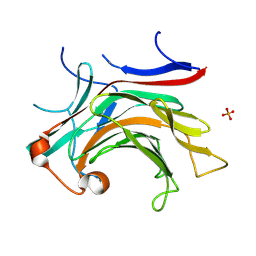 | | Crystal structure of CapGH16_3 enzyme retrieved from capybara gut metagenome | | Descriptor: | CALCIUM ION, Glycoside hydrolase family 16, PHOSPHATE ION | | Authors: | Vieira, P.S, Martins, M.P, Morais, M.A.B, Mandelli, F, Chinaglia, M, Lima, E.A, Murakami, M.T. | | Deposit date: | 2023-12-11 | | Release date: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | A functionally augmented carbohydrate utilization locus from herbivore gut microbiota fueled by dietary beta-glucans.
NPJ Biofilms Microbiomes, 10, 2024
|
|
8VA3
 
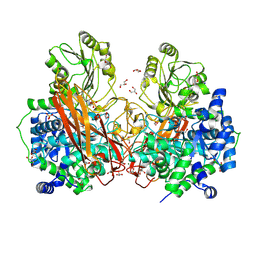 | | Crystal structure of CapGH3b enzyme retrieved from capybara gut metagenome | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Martins, M.P, Morais, M.A.B, Chinaglia, M, Mandelli, F, Lima, E.A, Murakami, M.T. | | Deposit date: | 2023-12-11 | | Release date: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A functionally augmented carbohydrate utilization locus from herbivore gut microbiota fueled by dietary beta-glucans.
NPJ Biofilms Microbiomes, 10, 2024
|
|
8Z2W
 
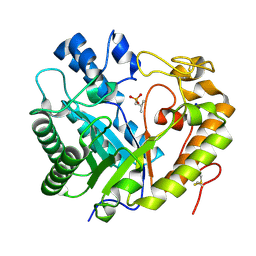 | | Crystal structure of apo- exo-beta-(1,3)-glucanase from Aspergillus oryzae (AoBgl) | | Descriptor: | 2-methylpropylphosphonic acid, Glucan 1,3-beta-glucosidase A, SODIUM ION | | Authors: | Banerjee, B, Kamale, C.K, Suryawanshi, A.B, Bhaumik, P. | | Deposit date: | 2024-04-13 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structures of Aspergillus oryzae exo-beta-(1,3)-glucanase reveal insights into oligosaccharide binding, recognition, and hydrolysis.
Febs Lett., 2024
|
|
5KPV
 
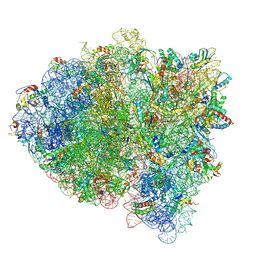 | | Structure of RelA bound to ribosome in presence of A/R tRNA (Structure II) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Loveland, A.B, Bah, E, Madireddy, R, Zhang, Y, Brilot, A.F, Grigorieff, N, Korostelev, A.A. | | Deposit date: | 2016-07-05 | | Release date: | 2016-09-28 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Ribosome•RelA structures reveal the mechanism of stringent response activation.
Elife, 5, 2016
|
|
1SI1
 
 | | Crystal Structure of Mannheimia haemolytica Ferric iron-Binding Protein A in an open conformation | | Descriptor: | FE (III) ION, iron binding protein FbpA | | Authors: | Shouldice, S.R, Skene, R.J, Dougan, D.R, Snell, G, McRee, D.E, Schryvers, A.B, Tari, L.W. | | Deposit date: | 2004-02-26 | | Release date: | 2004-06-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural basis for iron binding and release by a novel class of periplasmic iron-binding proteins found in gram-negative pathogens.
J.Bacteriol., 186, 2004
|
|
5KPW
 
 | | Structure of RelA bound to ribosome in presence of A/R tRNA (Structure III) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Loveland, A.B, Bah, E, Madireddy, R, Zhang, Y, Brilot, A.F, Grigorieff, N, Korostelev, A.A. | | Deposit date: | 2016-07-05 | | Release date: | 2016-09-28 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Ribosome•RelA structures reveal the mechanism of stringent response activation.
Elife, 5, 2016
|
|
5LAU
 
 | | Oceanobacillus iheyensis macrodomain mutant G37V with ADPR | | Descriptor: | GLYCEROL, MacroD-type macrodomain, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Gil-Ortiz, F, Zapata-Perez, R, Martinez, A.B, Juanhuix, J, Sanchez-Ferrer, A. | | Deposit date: | 2016-06-15 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural and functional analysis of Oceanobacillus iheyensis macrodomain reveals a network of waters involved in substrate binding and catalysis.
Open Biol, 7, 2017
|
|
6MGV
 
 | |
6MLM
 
 | | H7 HA0 in complex with Fv from H7.5 IgG | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain Fv of H7.5 Fab, Hemagglutinin HA1 chain, ... | | Authors: | Pallesen, J, Turner, H.L, Ward, A.B. | | Deposit date: | 2018-09-27 | | Release date: | 2019-02-20 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Potent anti-influenza H7 human monoclonal antibody induces separation of hemagglutinin receptor-binding head domains.
PLoS Biol., 17, 2019
|
|
1Q0U
 
 | |
5LBP
 
 | | Oceanobacillus iheyensis macrodomain mutant N30A | | Descriptor: | MacroD-type macrodomain, PHOSPHATE ION | | Authors: | Gil-Ortiz, F, Zapata-Perez, R, Martinez, A.B, Juanhuix, J, Sanchez-Ferrer, A. | | Deposit date: | 2016-06-16 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural and functional analysis of Oceanobacillus iheyensis macrodomain reveals a network of waters involved in substrate binding and catalysis.
Open Biol, 7, 2017
|
|
6MZC
 
 | | Human TFIID BC core | | Descriptor: | Transcription initiation factor TFIID subunit 10, Transcription initiation factor TFIID subunit 12, Transcription initiation factor TFIID subunit 2, ... | | Authors: | Patel, A.B, Louder, R.K, Greber, B.J, Grunberg, S, Luo, J, Fang, J, Liu, Y, Ranish, J, Hahn, S, Nogales, E. | | Deposit date: | 2018-11-05 | | Release date: | 2018-11-28 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structure of human TFIID and mechanism of TBP loading onto promoter DNA.
Science, 362, 2018
|
|
5KPS
 
 | | Structure of RelA bound to ribosome in absence of A/R tRNA (Structure I) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Loveland, A.B, Bah, E, Madireddy, R, Zhang, Y, Brilot, A.F, Grigorieff, N, Korostelev, A.A. | | Deposit date: | 2016-07-05 | | Release date: | 2016-09-28 | | Last modified: | 2019-11-20 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Ribosome•RelA structures reveal the mechanism of stringent response activation.
Elife, 5, 2016
|
|
6MGW
 
 | |
6MS3
 
 | | Crystal structure of the GH43 protein BlXynB mutant (K247S) from Bacillus licheniformis | | Descriptor: | CALCIUM ION, GLYCEROL, Glycoside Hydrolase Family 43, ... | | Authors: | Zanphorlin, L.M, Morais, M.A.B, Diogo, J.A, Murakami, M.T. | | Deposit date: | 2018-10-16 | | Release date: | 2019-04-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-guided design combined with evolutionary diversity led to the discovery of the xylose-releasing exo-xylanase activity in the glycoside hydrolase family 43.
Biotechnol. Bioeng., 116, 2019
|
|
6MZM
 
 | | Human TFIID bound to promoter DNA and TFIIA | | Descriptor: | SCP DNA (80-MER), TATA-box-binding protein, Transcription initiation factor IIA subunit 1, ... | | Authors: | Patel, A.B, Louder, R.K, Greber, B.J, Grunberg, S, Luo, J, Fang, J, Liu, Y, Ranish, J, Hahn, S, Nogales, E. | | Deposit date: | 2018-11-05 | | Release date: | 2018-11-28 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | Structure of human TFIID and mechanism of TBP loading onto promoter DNA.
Science, 362, 2018
|
|
6MZD
 
 | | Human TFIID Lobe A canonical | | Descriptor: | TATA-box-binding protein, Transcription initiation factor TFIID subunit 1, Transcription initiation factor TFIID subunit 10, ... | | Authors: | Patel, A.B, Louder, R.K, Greber, B.J, Grunberg, S, Luo, J, Fang, J, Liu, Y, Ranish, J, Hahn, S, Nogales, E. | | Deposit date: | 2018-11-05 | | Release date: | 2018-11-28 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (9.8 Å) | | Cite: | Structure of human TFIID and mechanism of TBP loading onto promoter DNA.
Science, 362, 2018
|
|
6MS2
 
 | | Crystal structure of the GH43 BlXynB protein from Bacillus licheniformis | | Descriptor: | CALCIUM ION, Glycoside Hydrolase Family 43 | | Authors: | Zanphorlin, L.M, Morais, M.A.B, Diogo, J.A, Murakami, M.T. | | Deposit date: | 2018-10-16 | | Release date: | 2019-04-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.494 Å) | | Cite: | Structure-guided design combined with evolutionary diversity led to the discovery of the xylose-releasing exo-xylanase activity in the glycoside hydrolase family 43.
Biotechnol. Bioeng., 116, 2019
|
|
5L9Q
 
 | | OCEANOBACILLUS IHEYENSIS MACRODOMAIN WITH ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MACROD-TYPE MACRODOMAIN, SULFATE ION | | Authors: | Gil-Ortiz, F, Zapata-Perez, R, Martinez, A.B, Juanhuix, J, Sanchez-Ferrer, A. | | Deposit date: | 2016-06-10 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and functional analysis of Oceanobacillus iheyensis macrodomain reveals a network of waters involved in substrate binding and catalysis.
Open Biol, 7, 2017
|
|
