8FYO
 
 | | MicroED structure of Proteinase K from lamellae milled from multiple plasma sources | | Descriptor: | CALCIUM ION, NITRATE ION, Proteinase K | | Authors: | Martynowycz, M.W, Shiriaeva, A, Clabbers, M.T.B, Nicolas, W.J, Weaver, S.J, Hattne, J, Gonen, T. | | Deposit date: | 2023-01-26 | | Release date: | 2023-05-24 | | Last modified: | 2024-10-23 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.39 Å) | | Cite: | A robust approach for MicroED sample preparation of lipidic cubic phase embedded membrane protein crystals.
Nat Commun, 14, 2023
|
|
6XOO
 
 | | DCN1 bound to DI-1859 | | Descriptor: | Lysozyme, DCN1-like protein 1 chimera, N-{(1S)-1-cyclohexyl-2-[(2-methylpropanoyl)amino]ethyl}-N~2~-propanoyl-3-[6-(propan-2-yl)-1,3-benzothiazol-2-yl]-L-alaninamide | | Authors: | Stuckey, J.A. | | Deposit date: | 2020-07-07 | | Release date: | 2021-06-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Selective inhibition of cullin 3 neddylation through covalent targeting DCN1 protects mice from acetaminophen-induced liver toxicity.
Nat Commun, 12, 2021
|
|
1D54
 
 | |
7A34
 
 | |
8GB6
 
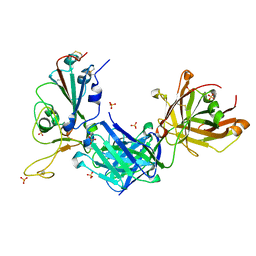 | |
6OUI
 
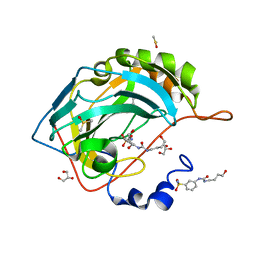 | |
7A2L
 
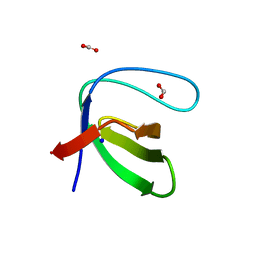 | |
4XY8
 
 | | Crystal Structure of the bromodomain of BRD9 in complex with a 2-amine-9H-purine ligand | | Descriptor: | 6-(5-bromo-2-methoxyphenyl)-9H-purin-2-amine, BROMIDE ION, Bromodomain-containing protein 9 | | Authors: | Picaud, S, von Delft, F, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Filippakopoulos, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-02-02 | | Release date: | 2015-03-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 9H-Purine Scaffold Reveals Induced-Fit Pocket Plasticity of the BRD9 Bromodomain.
J.Med.Chem., 58, 2015
|
|
6XOL
 
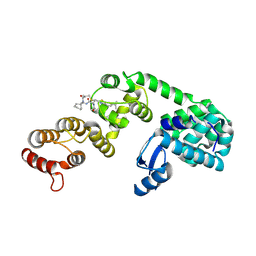 | | DCN1 bound to DI-1548 | | Descriptor: | Lysozyme, DCN1-like protein 1 chimera, N-{(1S)-1-cyclohexyl-2-[(2-methylpropanoyl)amino]ethyl}-N~2~-propanoyl-3-[6-(propan-2-yl)-1,3-benzothiazol-2-yl]-L-alaninamide | | Authors: | Stuckey, J.A. | | Deposit date: | 2020-07-07 | | Release date: | 2021-06-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Selective inhibition of cullin 3 neddylation through covalent targeting DCN1 protects mice from acetaminophen-induced liver toxicity.
Nat Commun, 12, 2021
|
|
8CO5
 
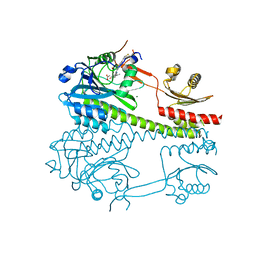 | |
2WFC
 
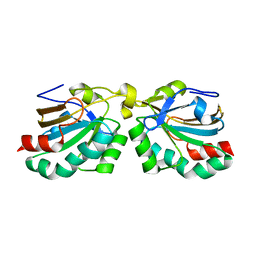 | |
8GKC
 
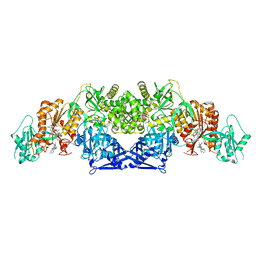 | |
6XA3
 
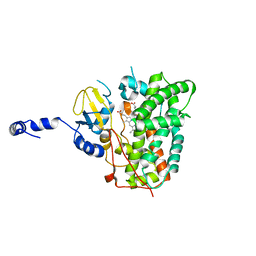 | | Structure of the ligand free P450 monooxygenase TamI | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, TamI | | Authors: | Newmister, S.A, Srivastava, K.R, Espinoza, R.V, Haatveit, K.C, Khatri, Y, Martini, R.M, Garcia-Borras, M, Podust, L.M, Houk, K.N, Sherman, D.H. | | Deposit date: | 2020-06-03 | | Release date: | 2021-06-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Molecular Basis of Iterative C─H Oxidation by TamI, a Multifunctional P450 monooxygenase from the Tirandamycin Biosynthetic Pathway.
Acs Catalysis, 10, 2020
|
|
8GLV
 
 | |
6S1E
 
 | | Structure of thaumatin determined at SwissFEL using native-SAD at 6.06 keV from all available diffraction patterns | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin-1 | | Authors: | Nass, K, Cheng, R, Vera, L, Mozzanica, A, Redford, S, Ozerov, D, Basu, S, James, D, Knopp, G, Cirelli, C, Martiel, I, Casadei, C, Weinert, T, Nogly, P, Skopintsev, P, Usov, I, Leonarski, F, Geng, T, Rappas, M, Dore, A.S, Cooke, R, Nasrollahi Shirazi, S, Dworkowski, F, Sharpe, M, Olieric, N, Steinmetz, M.O, Schertler, G, Abela, R, Patthey, L, Schmitt, B, Hennig, M, Standfuss, J, Wang, M, Milne, J.C. | | Deposit date: | 2019-06-18 | | Release date: | 2020-07-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Advances in long-wavelength native phasing at X-ray free-electron lasers.
Iucrj, 7, 2020
|
|
8TG5
 
 | | tRNA 2'-phosphotransferase (Tpt1) from Pyrococcus horikoshii in complex with branched 2'-PO4 RNA | | Descriptor: | (2S,3R,4R,5S)-2-(6-amino-9H-purin-9-yl)-4-{[(S)-{[(2R,3S,4R,5S)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]methoxy}(hydroxy)phosphoryl]oxy}-5-({[(S)-{[(2R,3R,4S,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-2-(hydroxymethyl)oxolan-3-yl]oxy}(hydroxy)phosphoryl]oxy}methyl)oxolan-3-yl dihydrogen phosphate (non-preferred name), CHLORIDE ION, POTASSIUM ION, ... | | Authors: | Jacewicz, A, Dantuluri, S, Shuman, S. | | Deposit date: | 2023-07-12 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for Tpt1-catalyzed 2'-PO 4 transfer from RNA and NADP(H) to NAD.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
1CQG
 
 | | HIGH RESOLUTION SOLUTION NMR STRUCTURE OF MIXED DISULFIDE INTERMEDIATE BETWEEN HUMAN THIOREDOXIN (C35A, C62A, C69A, C73A) MUTANT AND A 13 RESIDUE PEPTIDE COMPRISING ITS TARGET SITE IN HUMAN REF-1 (RESIDUES 59-71 OF THE P50 SUBUNIT OF NFKB), NMR, 31 STRUCTURES | | Descriptor: | REF-1 PEPTIDE, THIOREDOXIN | | Authors: | Clore, G.M, Qin, J, Gronenborn, A.M. | | Deposit date: | 1996-04-02 | | Release date: | 1996-08-01 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | The solution structure of human thioredoxin complexed with its target from Ref-1 reveals peptide chain reversal.
Structure, 4, 1996
|
|
5MCZ
 
 | | The structure of the mature HIV-1 CA hexameric lattice with curvature parameters: tilt=-1, twist=0 | | Descriptor: | Capsid protein p24 | | Authors: | Mattei, S, Glass, B, Hagen, W.J.H, Kraeusslich, H.-G, Briggs, J.A.G. | | Deposit date: | 2016-11-10 | | Release date: | 2016-12-28 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (8.2 Å) | | Cite: | The structure and flexibility of conical HIV-1 capsids determined within intact virions.
Science, 354, 2016
|
|
1CW8
 
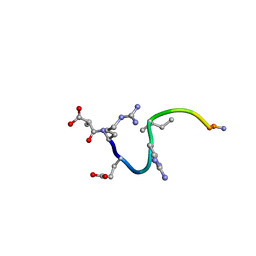 | | SOLUTION STRUCTURE OF THE ANALOGUE RETRO-INVERSO (mA-R)REGRIGGC IN CONTACT WITH THE MONOCLONAL ANTIBODY MAB 4X11, NMR, 6 STRUCTURES | | Descriptor: | HISTONE H3, METHYLMALONIC ACID | | Authors: | Phan Chan Du, A, Petit, M.C, Guichard, G, Briand, J.P, Muller, S, Cung, T. | | Deposit date: | 1999-08-26 | | Release date: | 1999-09-03 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Structure of antibody-bound peptides and retro-inverso analogues. A transferred nuclear Overhauser effect spectroscopy and molecular dynamics approach.
Biochemistry, 40, 2001
|
|
5MDB
 
 | | The structure of the mature HIV-1 CA hexameric lattice with curvature parameters: tilt=17, twist=-6 | | Descriptor: | Gag protein | | Authors: | Mattei, S, Glass, B, Hagen, W.J.H, Kraeusslich, H.-G, Briggs, J.A.G. | | Deposit date: | 2016-11-10 | | Release date: | 2016-12-28 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (8.4 Å) | | Cite: | The structure and flexibility of conical HIV-1 capsids determined within intact virions.
Science, 354, 2016
|
|
1CXF
 
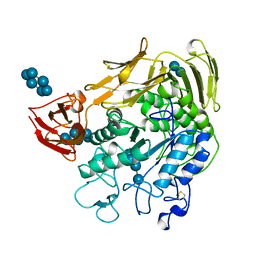 | | COMPLEX OF A (D229N/E257Q) DOUBLE MUTANT CGTASE FROM BACILLUS CIRCULANS STRAIN 251 WITH MALTOTETRAOSE AT 120 K AND PH 9.1 OBTAINED AFTER SOAKING THE CRYSTAL WITH ALPHA-CYCLODEXTRIN | | Descriptor: | CALCIUM ION, CYCLODEXTRIN GLYCOSYLTRANSFERASE, Cyclohexakis-(1-4)-(alpha-D-glucopyranose), ... | | Authors: | Knegtel, R.M.A, Strokopytov, B.V, Dijkstra, B.W. | | Deposit date: | 1995-07-28 | | Release date: | 1995-12-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystallographic studies of the interaction of cyclodextrin glycosyltransferase from Bacillus circulans strain 251 with natural substrates and products.
J.Biol.Chem., 270, 1995
|
|
4YKQ
 
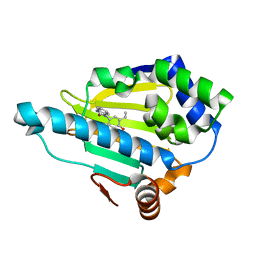 | | Heat Shock Protein 90 Bound to CS301 | | Descriptor: | 1-(5-ethyl-2,4-dihydroxyphenyl)-1,3-dihydro-2H-benzimidazol-2-one, Heat shock protein HSP 90-alpha | | Authors: | Kang, Y.N, Stuckey, J.A. | | Deposit date: | 2015-03-04 | | Release date: | 2016-03-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structure of Heat Shock Protein 90 Bound to CS301
To Be Published
|
|
1CTM
 
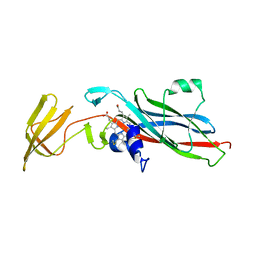 | | CRYSTAL STRUCTURE OF CHLOROPLAST CYTOCHROME F REVEALS A NOVEL CYTOCHROME FOLD AND UNEXPECTED HEME LIGATION | | Descriptor: | CYTOCHROME F, HEME C | | Authors: | Martinez, S.E, Huang, D, Szczepaniak, A, Cramer, W.A, Smith, J.L. | | Deposit date: | 1994-01-02 | | Release date: | 1994-05-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of chloroplast cytochrome f reveals a novel cytochrome fold and unexpected heme ligation.
Structure, 2, 1994
|
|
1D3W
 
 | | Crystal structure of ferredoxin 1 d15e mutant from azotobacter vinelandii at 1.7 angstrom resolution. | | Descriptor: | FE3-S4 CLUSTER, FERREDOXIN 1, IRON/SULFUR CLUSTER | | Authors: | Chen, K, Hirst, J, Camba, R, Bonagura, C.A, Stout, C.D, Burges, B.K, Armstrong, F.A. | | Deposit date: | 1999-10-01 | | Release date: | 1999-10-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Atomically defined mechanism for proton transfer to a buried redox centre in a protein.
Nature, 405, 2000
|
|
6XN8
 
 | | Crystal Structure of 2-hydroxyacyl CoA lyase (HACL) from Rhodospirillales bacterium URHD0017 | | Descriptor: | 2-hydroxyacyl-CoA lyase 1, 2-{3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-4-METHYL-2-OXO-2,3-DIHYDRO-1,3-THIAZOL-5-YL}ETHYL TRIHYDROGEN DIPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Miller, M.D, Xu, W, Olmos Jr, J.L, Chou, A, Clomburg, J.M, Gonzalez, R, Philips Jr, G.N. | | Deposit date: | 2020-07-02 | | Release date: | 2021-07-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of 2-hydroxyacyl CoA lyase (HACL) from Rhodospirillales bacterium URHD0017
To Be Published
|
|
