7UNO
 
 | | Thiol-disulfide oxidoreductase TsdA, C129S mutant, from Corynebacterium diphtheriae | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, TRIETHYLENE GLYCOL, ... | | Authors: | Osipiuk, J, Reardon-Robinson, M, Nguyen, M.T, Sanchez, B, Ton-That, H, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-04-11 | | Release date: | 2022-04-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Thiol-disulfide oxidoreductase in Corynebacterium diphtheriae
To Be Published
|
|
7RNX
 
 | | CLC-ec1 at pH 7.5 100mM Cl Swap | | Descriptor: | H(+)/Cl(-) exchange transporter ClcA | | Authors: | Fortea, E, Boudker, O, Accardi, A. | | Deposit date: | 2021-07-30 | | Release date: | 2022-11-16 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | Structural basis of common gate activation in CLC transporters
To Be Published
|
|
7RP6
 
 | |
7RO0
 
 | |
7RQ7
 
 | |
7RP5
 
 | | CLC-ec1 at pH 4.5 100mM Cl Turn | | Descriptor: | H(+)/Cl(-) exchange transporter ClcA | | Authors: | Fortea, E, Boudker, O, Accardi, A. | | Deposit date: | 2021-08-03 | | Release date: | 2022-11-16 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | Structural basis of common gate activation in CLC transporters
To Be Published
|
|
7PF1
 
 | | UVC treated Human apoferritin | | Descriptor: | CHLORIDE ION, Ferritin heavy chain, N-terminally processed, ... | | Authors: | Renault, L, Depelteau, J.S, Briegel, A. | | Deposit date: | 2021-08-11 | | Release date: | 2022-01-26 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.1 Å) | | Cite: | UVC inactivation of pathogenic samples suitable for cryo-EM analysis.
Commun Biol, 5, 2022
|
|
7PAG
 
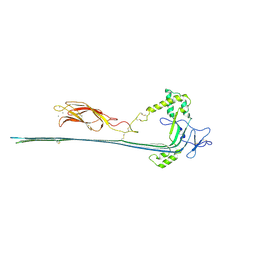 | | The pore conformation of lymphocyte perforin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Perforin-1 | | Authors: | Ivanova, M.E, Lukoyanova, N, Malhotra, S, Topf, M, Trapani, J.A, Voskoboinik, I, Saibil, H.R. | | Deposit date: | 2021-07-29 | | Release date: | 2022-02-16 | | Last modified: | 2022-02-23 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | The pore conformation of lymphocyte perforin.
Sci Adv, 8, 2022
|
|
7QXV
 
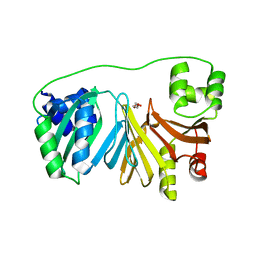 | | Crystal Structure of Haem-Binding Protein HemS Mutant F104A F199A, from Yersinia enterocolitica | | Descriptor: | DI(HYDROXYETHYL)ETHER, Hemin transport protein | | Authors: | Barker, P.D, Keith, A, Brear, P, Wales, D. | | Deposit date: | 2022-01-27 | | Release date: | 2023-02-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Crystal Structure of Haem-Binding Protein HemS Mutant F104A F199A, from Yersinia enterocolitica
To Be Published
|
|
7PW1
 
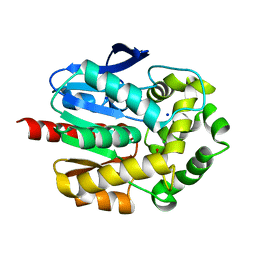 | | Crystal structure of ancestral haloalkane dehalogenase AncLinB-DmbA | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Haloalkane dehalogenase, ... | | Authors: | Mazur, A, Grinkevich, P, Prudnikova, T. | | Deposit date: | 2021-10-05 | | Release date: | 2022-04-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Analysis of the Ancestral Haloalkane Dehalogenase AncLinB-DmbA.
Int J Mol Sci, 22, 2021
|
|
7Q5C
 
 | | Crystal structure of OmpG in space group 96 | | Descriptor: | Outer membrane porin G, SODIUM ION, TETRAETHYLENE GLYCOL, ... | | Authors: | Nguyen, T.T.M, Khan, A.R, Barringer, R, McManus, J.J, Race, P.R. | | Deposit date: | 2021-11-03 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.717 Å) | | Cite: | Experimental phase diagrams to optimise OmpG
To Be Published
|
|
7S7X
 
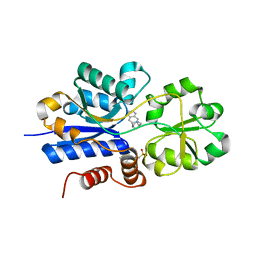 | | Crystal structure of iCytSnFR Cytisine Sensor precursor binding protein with varenicline bound | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, DI(HYDROXYETHYL)ETHER, VARENICLINE, ... | | Authors: | Fan, C, Nichols, N.L, Luebbert, L, Looger, L.L, Lester, H.A, Rees, D.C. | | Deposit date: | 2021-09-17 | | Release date: | 2021-10-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure, Function, and Application of Bacterial ABC Transporters
Ph.D.Thesis,California Institute of Technology, 2020
|
|
7S7W
 
 | | Crystal structure of iNicSnFR3a Nicotine Sensor precursor binding protein | | Descriptor: | 1,2-ETHANEDIOL, SODIUM ION, iNicSnFR 3.0 Fluorescent Nicotine Sensor precursor binding protein | | Authors: | Fan, C, Shivange, A.V, Looger, L.L, Lester, H.A, Rees, D.C. | | Deposit date: | 2021-09-17 | | Release date: | 2021-10-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structure, Function, and Application of Bacterial ABC Transporters
Ph.D.Thesis,California Institute of Technology, 2020
|
|
2GGO
 
 | | Crystal Structure of glucose-1-phosphate thymidylyltransferase from Sulfolobus tokodaii | | Descriptor: | 401aa long hypothetical glucose-1-phosphate thymidylyltransferase | | Authors: | Rajakannan, V, Mizushima, T, Suzuki, A, Masui, R, Kuramitsu, S, Yamane, T. | | Deposit date: | 2006-03-24 | | Release date: | 2007-03-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of glucose-1-phosphate thymidylyltransferase from
Sulfolobus tokodaii
To be published
|
|
6Y4I
 
 | |
6YI6
 
 | |
6YMS
 
 | |
6GMR
 
 | | pVHL:EloB:EloC in complex with (4-(1H-pyrrol-1-yl)phenyl)methanol | | Descriptor: | (4-pyrrol-1-ylphenyl)methanol, Elongin-B, Elongin-C, ... | | Authors: | Van Molle, I, Lucas, X, Ciulli, A. | | Deposit date: | 2018-05-28 | | Release date: | 2018-08-08 | | Last modified: | 2018-09-05 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Surface Probing by Fragment-Based Screening and Computational Methods Identifies Ligandable Pockets on the von Hippel-Lindau (VHL) E3 Ubiquitin Ligase.
J. Med. Chem., 61, 2018
|
|
6YN3
 
 | | Thrombin in complex with 4-hydroxybenzamide (j89) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-HYDROXYBENZAMIDE, DIMETHYL SULFOXIDE, ... | | Authors: | Scanlan, W, Heine, A, Klebe, G, Abazi, N. | | Deposit date: | 2020-04-10 | | Release date: | 2021-04-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Thrombin in complex with 4-hydroxybenzamide (j89)
To be published
|
|
6YO5
 
 | | Crystal structure of the M295F variant of Ssl1 | | Descriptor: | ALA-HIS-ALA, COPPER (II) ION, Copper oxidase, ... | | Authors: | Mielenbrink, S, Olbrich, A, Urlacher, V, Span, I. | | Deposit date: | 2020-04-14 | | Release date: | 2021-04-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Effect of the axial ligand mutation on spectral and structural properties of Ssl1 laccase
To Be Published
|
|
6YMP
 
 | | Thrombin in complex with 3-((5-(tert-butyl)isoxazol-3-yl)methyl)oxetan-3-amine (j54) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-[(5-tert-butyl-1,2-oxazol-3-yl)methyl]oxetan-3-amine, DIMETHYL SULFOXIDE, ... | | Authors: | Scanlan, W, Heine, A, Klebe, G, Abazi, N. | | Deposit date: | 2020-04-09 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Thrombin in complex with 3-((5-(tert-butyl)isoxazol-3-yl)methyl)oxetan-3-amine (j54)
To be published
|
|
6YMR
 
 | |
6YQV
 
 | | Thrombin in complex with 5-chlorothiophene-2-sulfonamide (j94) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-chloranylthiophene-2-sulfonamide, DIMETHYL SULFOXIDE, ... | | Authors: | Scanlan, W, Heine, A, Klebe, G, Abazi, N. | | Deposit date: | 2020-04-18 | | Release date: | 2021-04-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Thrombin in complex with 5-chlorothiophene-2-sulfonamide (j94)
To be published
|
|
6H5G
 
 | | Crystal structure of DHQ1 from Salmonella typhi covalently modified by ligand 3 | | Descriptor: | (1~{R},3~{S},4~{R},5~{R})-3-methyl-4,5-bis(hydroxyl)cyclohexane-1-carboxylic acid, 3-dehydroquinate dehydratase | | Authors: | Sanz-Gaitero, M, Maneiro, M, Lence, E, Otero, J.M, Thompson, P, Hawkins, A.R, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2018-07-24 | | Release date: | 2019-07-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Hydroxylammonium Derivatives for Selective Active-site Lysine Modification in the Anti-virulence Bacterial Target DHQ1 Enzyme.
Org Chem Front, 6, 2019
|
|
6Y4A
 
 | |
