6RH0
 
 | | Revisiting pH-gated conformational switch. Complex HK853-RR468 pH 5.5 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Response regulator, ... | | Authors: | Mideros-Mora, C, Casino, P, Marina, A. | | Deposit date: | 2019-04-18 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Revisiting the pH-gated conformational switch on the activities of HisKA-family histidine kinases.
Nat Commun, 11, 2020
|
|
5K4G
 
 | |
5K50
 
 | | Three-dimensional structure of L-threonine 3-dehydrogenase from Trypanosoma brucei bound to NAD+ and L-allo-threonine refined to 2.23 angstroms | | Descriptor: | ACETATE ION, ALLO-THREONINE, GLYCEROL, ... | | Authors: | Adjogatse, E.A, Erskine, P.T, Cooper, J.B. | | Deposit date: | 2016-05-22 | | Release date: | 2017-11-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structure and function of L-threonine-3-dehydrogenase from the parasitic protozoan Trypanosoma brucei revealed by X-ray crystallography and geometric simulations.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
6EIY
 
 | | Crystal structure of KDM5B in complex with KDOPZ000034a. | | Descriptor: | 1,2-ETHANEDIOL, 2-chloranyl-~{N}-[2-[4-(3-cyano-7-oxidanylidene-6-propan-2-yl-4~{H}-pyrazolo[1,5-a]pyrimidin-5-yl)pyrazol-1-yl]ethyl]ethanamide, DIMETHYL SULFOXIDE, ... | | Authors: | Srikannathasan, V, Newman, J.A, Szykowska, A, Wright, M, Ruda, G.F, Vazquez-Rodriguez, S.A, Kupinska, K, Strain-Damerell, C, Burgess-Brown, N.A, Arrowsmith, C.H, Edwards, A, Bountra, C, Oppermann, U, Huber, K, von Delft, F. | | Deposit date: | 2017-09-19 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of KDM5B in complex with KDOPZ000034a.
to be published
|
|
7ASD
 
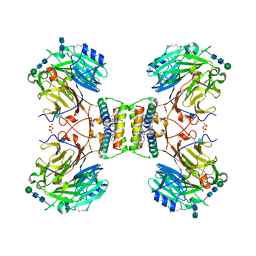 | | Structure of native royal jelly filaments | | Descriptor: | (3beta,14beta,17alpha)-ergosta-5,24(28)-dien-3-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Mattei, S, Ban, A, Picenoni, A, Leibundgut, M, Glockshuber, R, Boehringer, D. | | Deposit date: | 2020-10-27 | | Release date: | 2020-12-30 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of native glycolipoprotein filaments in honeybee royal jelly.
Nat Commun, 11, 2020
|
|
8JCQ
 
 | |
5K68
 
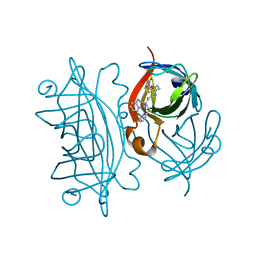 | | Designed Artificial Cupredoxins | | Descriptor: | Streptavidin, [CuII(biot-bu-dpea)]2+ | | Authors: | Mann, S.I, Heinisch, T, Weitz, A.C, Hendrich, M.R, Ward, T.R, Borovik, A.S. | | Deposit date: | 2016-05-24 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Modular Artificial Cupredoxins.
J.Am.Chem.Soc., 138, 2016
|
|
5GJG
 
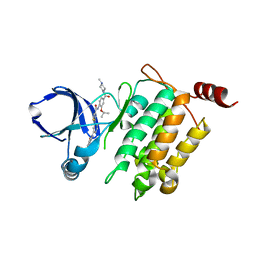 | | Crystal structure of human TAK1/TAB1 fusion protein in complex with ligand 4 | | Descriptor: | N-(2-isopropoxy-4-(4-methylpiperazine-1-carbonyl)phenyl)-2-(3-(phenylcarbamoyl)phenyl)thiazole-4-carboxamide, TAK1 kinase - TAB1 chimera fusion protein | | Authors: | Irie, M, Nakamura, M, Fukami, T.A, Matsuura, T, Morishima, K. | | Deposit date: | 2016-06-29 | | Release date: | 2016-11-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Development of a Method for Converting a TAK1 Type I Inhibitor into a Type II or c-Helix-Out Inhibitor by Structure-Based Drug Design (SBDD)
Chem.Pharm.Bull., 64, 2016
|
|
6IV3
 
 | |
8JCS
 
 | |
8UP3
 
 | | Structure of atypical asparaginase from Rhodospirillum rubrum (mutant Y21F) | | Descriptor: | 1,2-ETHANEDIOL, ASPARTIC ACID, Asparaginase, ... | | Authors: | Lubkowski, J, Wlodawer, A, Zhang, D. | | Deposit date: | 2023-10-20 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | RrA, an enzyme from Rhodospirillum rubrum, is a prototype of a new family of short-chain L-asparaginases.
Protein Sci., 33, 2024
|
|
6RIH
 
 | | Crystal structure of PHGDH in complex with compound 9 | | Descriptor: | D-3-phosphoglycerate dehydrogenase, SULFATE ION, ~{N}-cyclopropyl-2-methyl-5-phenyl-pyrazole-3-carboxamide | | Authors: | Bader, G, Wolkerstorfer, B, Zoephel, A. | | Deposit date: | 2019-04-24 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Intracellular Trapping of the Selective Phosphoglycerate Dehydrogenase (PHGDH) InhibitorBI-4924Disrupts Serine Biosynthesis.
J.Med.Chem., 62, 2019
|
|
8UPC
 
 | | Structure of atypical asparaginase from Rhodospirillum rubrum (mutant K158M) | | Descriptor: | Asparaginase, CHLORIDE ION, GLYCEROL | | Authors: | Lubkowski, J, Wlodawer, A, Zhang, D. | | Deposit date: | 2023-10-22 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | RrA, an enzyme from Rhodospirillum rubrum, is a prototype of a new family of short-chain L-asparaginases.
Protein Sci., 33, 2024
|
|
4R9X
 
 | | Crystal Structure of Putative Copper Homeostasis Protein CutC from Bacillus anthracis | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Copper homeostasis protein CutC, ... | | Authors: | Kim, Y, Zhou, M, Makowska-Grzyska, M, Grimshaw, S, Anderson, W.F, Joachimiak, A, CSGID, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-09-08 | | Release date: | 2014-09-17 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.8515 Å) | | Cite: | Crystal Structure of Putative Copper Homeostasis Protein CutC
from Bacillus anthracis
To be Published, 2014
|
|
8UP6
 
 | | Structure of atypical asparaginase from Rhodospirillum rubrum (mutant K19A) in complex with L-Asp | | Descriptor: | ASPARTIC ACID, Asparaginase, TETRAETHYLENE GLYCOL | | Authors: | Lubkowski, J, Wlodawer, A, Zhang, D. | | Deposit date: | 2023-10-21 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | RrA, an enzyme from Rhodospirillum rubrum, is a prototype of a new family of short-chain L-asparaginases.
Protein Sci., 33, 2024
|
|
8EAG
 
 | | SsoMCM hexamer bound to Mg/ADP-BeFx and 12-mer oligo-dT. Class 2 | | Descriptor: | 12-mer oligo-dT, MAGNESIUM ION, Minichromosome maintenance protein MCM, ... | | Authors: | Meagher, M, Myasnikov, A, Enemark, E.J. | | Deposit date: | 2022-08-29 | | Release date: | 2022-12-14 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Two Distinct Modes of DNA Binding by an MCM Helicase Enable DNA Translocation.
Int J Mol Sci, 23, 2022
|
|
6RIC
 
 | | Structure of the core Vaccinia Virus DNA-dependent RNA polymerase complex | | Descriptor: | DNA-dependent RNA polymerase subunit rpo132, DNA-dependent RNA polymerase subunit rpo147, DNA-dependent RNA polymerase subunit rpo18, ... | | Authors: | Grimm, C, Hillen, H.S, Bedenk, K, Bartuli, J, Neyer, S, Zhang, Q, Huettenhofer, A, Erlacher, M, Dienemann, C, Schlosser, A, Urlaub, H, Boettcher, B, Szalay, A, Cramer, P, Fischer, U. | | Deposit date: | 2019-04-23 | | Release date: | 2019-12-18 | | Last modified: | 2019-12-25 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural Basis of Poxvirus Transcription: Transcribing and Capping Vaccinia Complexes.
Cell, 179, 2019
|
|
5X6Y
 
 | |
6E6K
 
 | | Crystal structure of human cellular retinol-binding protein 4 in complex with abnormal-cannabidiol (abn-CBD) | | Descriptor: | (1'R,2'R)-5'-methyl-6-pentyl-2'-(prop-1-en-2-yl)-1',2',3',4'-tetrahydro[1,1'-biphenyl]-2,4-diol, Retinoid-binding protein 7 | | Authors: | Silvaroli, J.A, Banerjee, S, Kiser, P.D, Golczak, M. | | Deposit date: | 2018-07-25 | | Release date: | 2019-02-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Abnormal Cannabidiol Modulates Vitamin A Metabolism by Acting as a Competitive Inhibitor of CRBP1.
Acs Chem.Biol., 14, 2019
|
|
6ALS
 
 | | Solution structure of a DNA dodecamer with 5-methylcytosine at the 3rd and 9th position and 8-oxoguanine at the 4th position | | Descriptor: | DNA (5'-D(*(DC5)P*GP*(DMC)P*(8OG)P*AP*AP*TP*TP*(DMC)P*GP*CP*(DG3))-3') | | Authors: | Gruber, D.R, Shernyukov, A.V, Endutkin, A.V, Bagryanskaya, E.G, Zharkov, D.O, Smirnov, S.L. | | Deposit date: | 2017-08-08 | | Release date: | 2017-09-06 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Oxidative damage to epigenetically methylated sites affects DNA stability, dynamics and enzymatic demethylation.
Nucleic Acids Res., 46, 2018
|
|
7M41
 
 | | Structure of TIM-3 in complex with N-(4-(8-chloro-2-methyl-5-oxo-5,6-dihydro-[1,2,4]traizolo[1,5-c]quinazolin-9-yl)-3-methylphenyl)-1H-imidazole-2-sulfonamide (compound 38) | | Descriptor: | CALCIUM ION, Hepatitis A virus cellular receptor 2, N-{4-[(4S,10aP)-8-chloro-2-methyl-5-oxo-5,6-dihydro[1,2,4]triazolo[1,5-c]quinazolin-9-yl]-3-methylphenyl}-1H-imidazole-2-sulfonamide | | Authors: | Rietz, T.A. | | Deposit date: | 2021-03-19 | | Release date: | 2021-10-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Fragment-Based Discovery of Small Molecules Bound to T-Cell Immunoglobulin and Mucin Domain-Containing Molecule 3 (TIM-3).
J.Med.Chem., 64, 2021
|
|
5KAR
 
 | | Murine acid sphingomyelinase-like phosphodiesterase 3b (SMPDL3B) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Acid sphingomyelinase-like phosphodiesterase 3b, ... | | Authors: | Gorelik, A, Illes, K, Heinz, L.X, Superti-Furga, G, Nagar, B. | | Deposit date: | 2016-06-02 | | Release date: | 2016-10-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.142 Å) | | Cite: | Crystal Structure of the Acid Sphingomyelinase-like Phosphodiesterase SMPDL3B Provides Insights into Determinants of Substrate Specificity.
J.Biol.Chem., 291, 2016
|
|
6I37
 
 | | Crystal structure of nv1Pizza6-AYW, a circularly permuted designer protein | | Descriptor: | SULFATE ION, nv1Pizza6-AYW | | Authors: | Mylemans, B, Noguchi, H, Deridder, E, Voet, A.R.D. | | Deposit date: | 2018-11-05 | | Release date: | 2019-11-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Influence of circular permutations on the structure and stability of a six-fold circular symmetric designer protein.
Protein Sci., 2020
|
|
5KB0
 
 | | Crystal Structure of a Tris-thiolate Pb(II) Complex in a de Novo Three-stranded Coiled Coil Peptide | | Descriptor: | CHLORIDE ION, LEAD (II) ION, Pb(II)Zn(II)(GRAND Coil Ser-L16CL30H)3+, ... | | Authors: | Ruckthong, L, Zastrow, M.L, Stuckey, J.A, Pecoraro, V.L. | | Deposit date: | 2016-06-02 | | Release date: | 2016-08-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | A Crystallographic Examination of Predisposition versus Preorganization in de Novo Designed Metalloproteins.
J.Am.Chem.Soc., 138, 2016
|
|
6I3H
 
 | | Crystal structure of influenza A virus M1 N-terminal domain (G18A mutation) | | Descriptor: | Matrix protein 1, PHOSPHATE ION | | Authors: | Miyake, Y, Keusch, J.J, Decamps, L, Ho-Xuan, H, Iketani, S, Gut, H, Kutay, U, Helenius, A, Yamauchi, Y. | | Deposit date: | 2018-11-06 | | Release date: | 2019-09-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Influenza virus uses transportin 1 for vRNP debundling during cell entry.
Nat Microbiol, 4, 2019
|
|
