7QD0
 
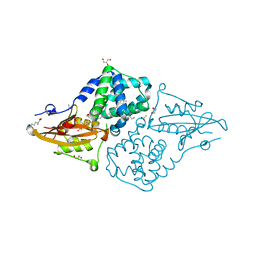 | | Structure of the orange carotenoid protein from Planktothrix agardhii binding echinenone in the C2 space group | | Descriptor: | ACETATE ION, ARGININE, GLYCEROL, ... | | Authors: | Andreeva, E.A, Hartmann, E, Schlichting, I, Colletier, J.-P. | | Deposit date: | 2021-11-25 | | Release date: | 2022-07-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-function-dynamics relationships in the peculiar Planktothrix PCC7805 OCP1: Impact of his-tagging and carotenoid type.
Biochim Biophys Acta Bioenerg, 1863, 2022
|
|
7QCZ
 
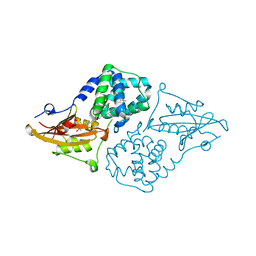 | | Structure of the orange carotenoid protein from Planktothrix agardhii binding canthaxanthin in the C2 space group | | Descriptor: | Orange carotenoid-binding protein, beta,beta-carotene-4,4'-dione | | Authors: | Andreeva, E.A, Hartmann, E, Schlichting, I, Colletier, J.-P. | | Deposit date: | 2021-11-25 | | Release date: | 2022-07-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-function-dynamics relationships in the peculiar Planktothrix PCC7805 OCP1: Impact of his-tagging and carotenoid type.
Biochim Biophys Acta Bioenerg, 1863, 2022
|
|
4XEA
 
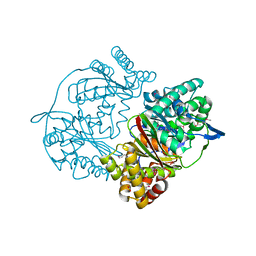 | | Crystal structure of putative M16-like peptidase from Alicyclobacillus acidocaldarius | | Descriptor: | ACETATE ION, GLYCEROL, NICKEL (II) ION, ... | | Authors: | Michalska, K, Tesar, C, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-12-23 | | Release date: | 2015-03-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of putative M16-like peptidase from Alicyclobacillus acidocaldarius
To Be Published
|
|
8PJB
 
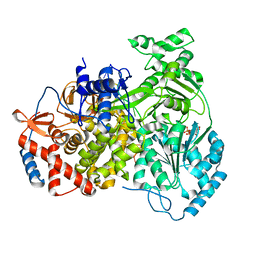 | |
8CZA
 
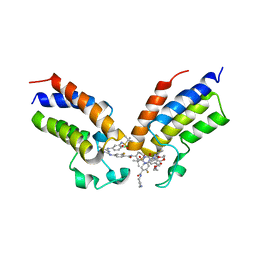 | | Crystal structure of the first bromodomain (BD1) of human BRDT bound to GXH-IV-075 | | Descriptor: | 4-[(4-{4-chloro-3-[(2-methylpropane-2-sulfonyl)amino]anilino}-5-methylpyrimidin-2-yl)amino]-2-fluoro-N-[1-(14-{3-[(2-{3-fluoro-4-[(piperidin-4-yl)carbamoyl]anilino}-5-methylpyrimidin-4-yl)amino]-5-[(2-methylpropane-2-sulfonyl)amino]phenyl}-14-oxo-4,7,10-trioxa-13-azatetradecanan-1-oyl)piperidin-4-yl]benzamide, Bromodomain testis-specific protein, SODIUM ION | | Authors: | Chan, A, Schonbrunn, E. | | Deposit date: | 2022-05-24 | | Release date: | 2022-08-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Bivalent BET Bromodomain Inhibitors Confer Increased Potency and Selectivity for BRDT via Protein Conformational Plasticity.
J.Med.Chem., 65, 2022
|
|
5HOU
 
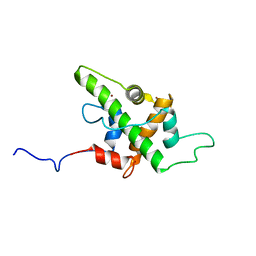 | | Solution Structure of p53TAD-TAZ1 | | Descriptor: | Cellular tumor antigen p53,CREB-binding protein fusion protein, ZINC ION | | Authors: | Krois, A.S, Ferreon, J.C, Martinez-Yamout, M.A, Dyson, H.J, Wright, P.E. | | Deposit date: | 2016-01-19 | | Release date: | 2016-03-16 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Recognition of the disordered p53 transactivation domain by the transcriptional adapter zinc finger domains of CREB-binding protein.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
8S8Y
 
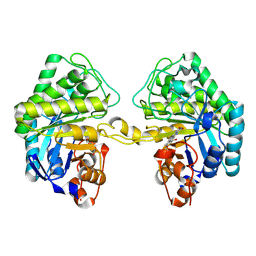 | | OPR3 variant R283E in its dimeric form | | Descriptor: | 12-oxophytodienoate reductase 3, FLAVIN MONONUCLEOTIDE | | Authors: | Bijelic, A, Macheroux, P, Kerschbaumer, B. | | Deposit date: | 2024-03-07 | | Release date: | 2024-08-14 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Analysis of homodimer formation in 12-oxophytodienoate reductase 3 in solutio and crystallo challenges the physiological role of the dimer.
Sci Rep, 14, 2024
|
|
3IR3
 
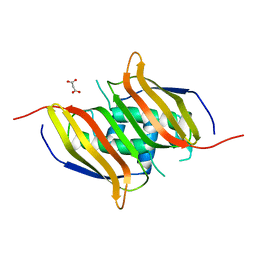 | | Crystal structure of human 3-hydroxyacyl-thioester dehydratase 2 (HTD2) | | Descriptor: | 3-hydroxyacyl-thioester dehydratase 2, MALONATE ION | | Authors: | Ugochukwu, E, Cocking, R, Burgess-Brown, N, Pilka, E, Muniz, J, Krojer, T, Chaikuad, A, Gileadi, O, von Delft, F, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A, Kavanagh, K.L, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-08-21 | | Release date: | 2009-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Crystal structure of human 3-hydroxyacyl-thioester dehydratase 2 (HTD2)
To be Published
|
|
6NXS
 
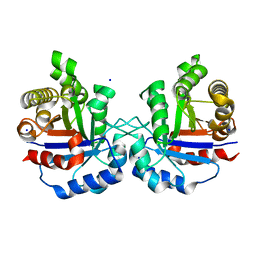 | | Crystal structure of Arabidopsis thaliana cytosolic triosephosphate isomerase C218Y mutant | | Descriptor: | SODIUM ION, Triosephosphate isomerase, cytosolic | | Authors: | Jimenez-Sandoval, P, Diaz-Quezada, C, Torres-Larios, A, Brieba, L.G. | | Deposit date: | 2019-02-09 | | Release date: | 2020-02-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Structural basis for the modulation of plant cytosolic triosephosphate isomerase activity by mimicry of redox-based modifications.
Plant J., 99, 2019
|
|
6NY1
 
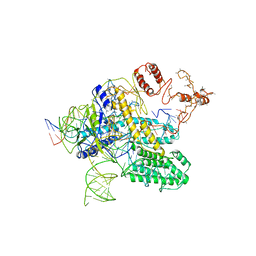 | | CasX-gRNA-DNA(30bp) State II | | Descriptor: | CasX, DNA Non-target strand, DNA target strand, ... | | Authors: | Liu, J.J, Orlova, N, Nogales, E, Doudna, J.A. | | Deposit date: | 2019-02-10 | | Release date: | 2019-02-27 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | CasX enzymes comprise a distinct family of RNA-guided genome editors.
Nature, 566, 2019
|
|
1YPK
 
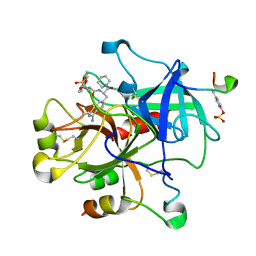 | | Thrombin Inhibitor Complex | | Descriptor: | 10-mer peptide from Acety-Hirudin(54-65) sulfated, Prothrombin heavy chain, Prothrombin light chain, ... | | Authors: | Czodrowski, P, Sotriffer, C, Fokkens, J, Heine, A, Klebe, G. | | Deposit date: | 2005-01-31 | | Release date: | 2006-01-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | A Simple Protocol To Estimate Differences in Protein Binding Affinity for Enantiomers without Prior Resolution of Racemates
Angew.Chem.Int.Ed.Engl., 45, 2006
|
|
5LJ8
 
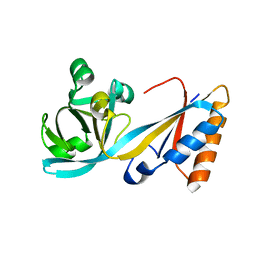 | | Structure of the E. coli MacB periplasmic domain (P21) | | Descriptor: | Macrolide export ATP-binding/permease protein MacB | | Authors: | Crow, A. | | Deposit date: | 2016-07-18 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure and mechanotransmission mechanism of the MacB ABC transporter superfamily.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5LMX
 
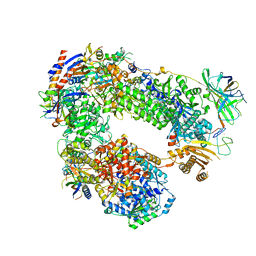 | | Monomeric RNA polymerase I at 4.9 A resolution | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | Authors: | Torreira, E, Louro, J.A, Gil-Carton, D, Gallego, O, Calvo, O, Fernandez-Tornero, C. | | Deposit date: | 2016-08-02 | | Release date: | 2017-03-22 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | The dynamic assembly of distinct RNA polymerase I complexes modulates rDNA transcription.
Elife, 6, 2017
|
|
6I0N
 
 | |
5LER
 
 | | Structure of the bacterial sex F pilus (13.2 Angstrom rise) | | Descriptor: | Pilin, [(2~{S})-3-[[(2~{R})-2,3-bis(oxidanyl)propoxy]-oxidanyl-phosphoryl]oxy-2-hexadec-9-enoyloxy-propyl] hexadecanoate | | Authors: | Costa, T.R.D, Ilangovan, I, Ukleja, M, Redzej, A, Santini, J.M, Smith, T.K, Egelman, E.H, Waksman, G. | | Deposit date: | 2016-06-30 | | Release date: | 2016-11-02 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Structure of the Bacterial Sex F Pilus Reveals an Assembly of a Stoichiometric Protein-Phospholipid Complex.
Cell, 166, 2016
|
|
6NX8
 
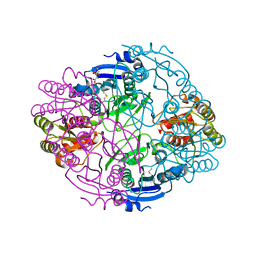 | |
4Y0S
 
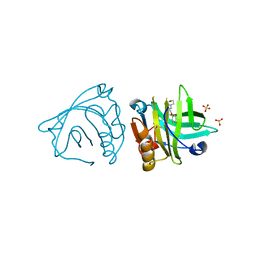 | | Goat beta-lactoglobulin complex with pramocaine (GLG-PRM) | | Descriptor: | Beta-lactoglobulin, Pramocaine, SULFATE ION | | Authors: | Loch, J.I, Bonarek, P, Polit, A, Jablonski, M, Czub, M, Ye, X, Lewinski, K. | | Deposit date: | 2015-02-06 | | Release date: | 2015-07-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | beta-Lactoglobulin interactions with local anaesthetic drugs - Crystallographic and calorimetric studies.
Int.J.Biol.Macromol., 80, 2015
|
|
6HXX
 
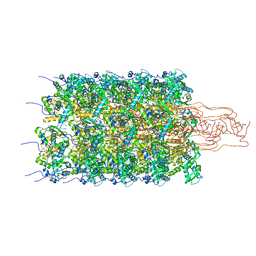 | | Potato virus Y | | Descriptor: | Coat protein, RNA (5'-R(P*UP*UP*UP*UP*U)-3') | | Authors: | Podobnik, M, Kezar, A, Novacek, J, Polak, M. | | Deposit date: | 2018-10-18 | | Release date: | 2019-08-07 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for the multitasking nature of the potato virus Y coat protein.
Sci Adv, 5, 2019
|
|
1YUD
 
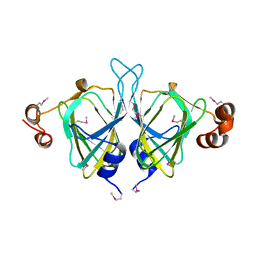 | | X-ray Crystal Structure of Protein SO0799 from Shewanella oneidensis. Northeast Structural Genomics Consortium Target SoR12. | | Descriptor: | hypothetical protein SO0799 | | Authors: | Kuzin, A.P, Vorobiev, S, Chen, Y, Forouhar, F, Acton, T, Ma, L.-C, Xiao, R, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-02-14 | | Release date: | 2005-03-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of hypothetical protein SO0799 from Shewanella oneidensis
To be Published
|
|
7LR5
 
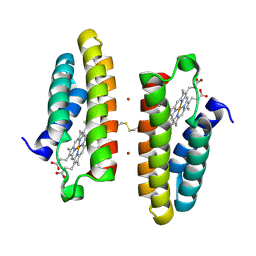 | |
5EV9
 
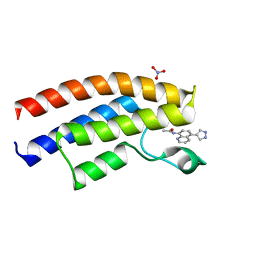 | | Crystal structure of the human BRPF1 bromodomain in complex with SEED15 | | Descriptor: | NITRATE ION, Peregrin, ~{N}-[5-(1~{H}-pyrazol-4-yl)quinolin-8-yl]ethanamide | | Authors: | Zhu, J, Caflisch, A. | | Deposit date: | 2015-11-19 | | Release date: | 2016-06-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Twenty Crystal Structures of Bromodomain and PHD Finger Containing Protein 1 (BRPF1)/Ligand Complexes Reveal Conserved Binding Motifs and Rare Interactions.
J.Med.Chem., 59, 2016
|
|
7LRR
 
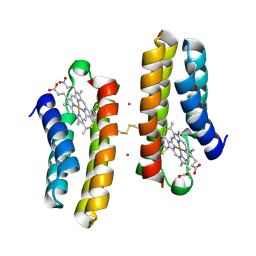 | |
7QOA
 
 | | Structure of CodB, a cytosine transporter in an outward-facing conformation | | Descriptor: | 2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 6-AMINOPYRIMIDIN-2(1H)-ONE, Cytosine permease, ... | | Authors: | Hatton, C.E, Cameron, A.D. | | Deposit date: | 2021-12-23 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of cytosine transport protein CodB provides insight into nucleobase-cation symporter 1 mechanism.
Embo J., 41, 2022
|
|
6HZL
 
 | | Crystal structure of redox-inhibited phosphoribulokinase from Synechococcus sp. (strain PCC 6301), osmate derivative | | Descriptor: | OSMIUM ION, Phosphoribulokinase | | Authors: | Wilson, R.H, Bracher, A, Hartl, F.U, Hayer-Hartl, M. | | Deposit date: | 2018-10-23 | | Release date: | 2019-03-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Crystal structure of phosphoribulokinase from Synechococcus sp. strain PCC 6301.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
7QOR
 
 | | Structure of beta-lactamase TEM-171 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, ... | | Authors: | Hakanpaa, J, Petrova, T, Samygina, V.R, Chojnowski, G, Lamzin, V, Egorov, A.M. | | Deposit date: | 2021-12-28 | | Release date: | 2022-07-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Crystal structures of the molecular class A beta-lactamase TEM-171 and its complexes with tazobactam.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
