8P78
 
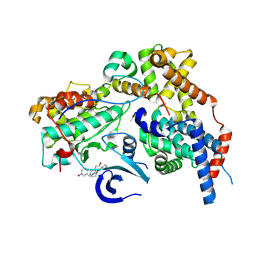 | | Cryo-EM structure of CAK in complex with inhibitor dinaciclib | | Descriptor: | 3-[({3-ethyl-5-[(2S)-2-(2-hydroxyethyl)piperidin-1-yl]pyrazolo[1,5-a]pyrimidin-7-yl}amino)methyl]-1-hydroxypyridinium, CDK-activating kinase assembly factor MAT1, Cyclin-H, ... | | Authors: | Cushing, V.I, Koh, A.F, Feng, J, Jurgaityte, K, Bahl, A.K, Ali, S, Kotecha, A, Greber, B.J. | | Deposit date: | 2023-05-30 | | Release date: | 2024-03-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (1.9 Å) | | Cite: | High-resolution cryo-EM of the human CDK-activating kinase for structure-based drug design.
Nat Commun, 15, 2024
|
|
8P79
 
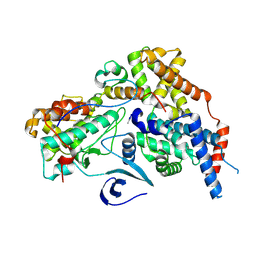 | | Cryo-EM structure of CAK with averaged inhibitor density | | Descriptor: | CDK-activating kinase assembly factor MAT1, Cyclin-H, Cyclin-dependent kinase 7 | | Authors: | Cushing, V.I, Koh, A.F, Feng, J, Jurgaityte, K, Bahl, A.K, Ali, S, Kotecha, A, Greber, B.J. | | Deposit date: | 2023-05-30 | | Release date: | 2024-03-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (1.7 Å) | | Cite: | High-resolution cryo-EM of the human CDK-activating kinase for structure-based drug design.
Nat Commun, 15, 2024
|
|
6Y45
 
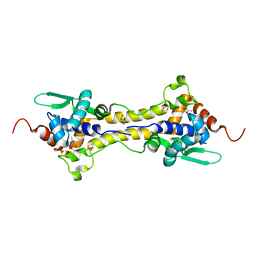 | | Crystal Structure of the H33A variant of RsrR | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Rohac, R, Volbeda, A, Fontecilla-Camps, J.C. | | Deposit date: | 2020-02-19 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Electron and Proton Transfers Modulate DNA Binding by the Transcription Regulator RsrR.
J.Am.Chem.Soc., 142, 2020
|
|
4XND
 
 | |
8TJ7
 
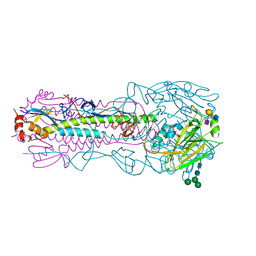 | | CRYSTAL STRUCTURE OF THE A/Shandong/9/1993(H3N2) INFLUENZA VIRUS HEMAGGLUTININ WITH HUMAN RECEPTOR ANALOG 6'-SLNLN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1 chain, ... | | Authors: | Wu, N.C, Zhu, X, Wilson, I.A. | | Deposit date: | 2023-07-20 | | Release date: | 2024-02-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Evolution of human H3N2 influenza virus receptor specificity has substantially expanded the receptor-binding domain site.
Cell Host Microbe, 32, 2024
|
|
8TJB
 
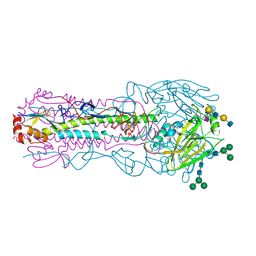 | | CRYSTAL STRUCTURE OF THE A/Texas/73/2017(H3N2) INFLUENZA VIRUS HEMAGGLUTININ WITH HUMAN RECEPTOR ANALOG 6'-SLNLN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1 chain, ... | | Authors: | Wu, N.C, Zhu, X, Wilson, I.A. | | Deposit date: | 2023-07-20 | | Release date: | 2024-02-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Evolution of human H3N2 influenza virus receptor specificity has substantially expanded the receptor-binding domain site.
Cell Host Microbe, 32, 2024
|
|
6ME3
 
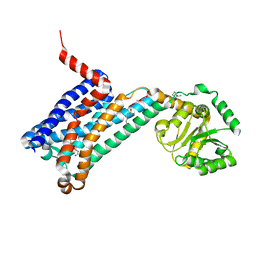 | | XFEL crystal structure of human melatonin receptor MT1 in complex with 2-phenylmelatonin | | Descriptor: | DI(HYDROXYETHYL)ETHER, N-[2-(5-methoxy-2-phenyl-1H-indol-3-yl)ethyl]acetamide, OLEIC ACID, ... | | Authors: | Stauch, B, Johansson, L.C, McCorvy, J.D, Patel, N, Han, G.W, Gati, C, Batyuk, A, Ishchenko, A, Brehm, W, White, T.A, Michaelian, N, Madsen, C, Zhu, L, Grant, T.D, Grandner, J.M, Olsen, R.H.J, Tribo, A.R, Weierstall, U, Roth, B.L, Katritch, V, Liu, W, Cherezov, V. | | Deposit date: | 2018-09-05 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of ligand recognition at the human MT1melatonin receptor.
Nature, 569, 2019
|
|
5LM6
 
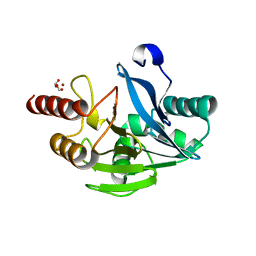 | | VIM-2 metallo-beta-lactamase in complex with 2-(3-fluoro-4-hydroxyphenyl)-3-oxoisoindoline-4-carboxylic acid (compound 35) | | Descriptor: | 2-(3-fluoranyl-4-oxidanyl-phenyl)-3-oxidanylidene-1~{H}-isoindole-4-carboxylic acid, FORMIC ACID, Metallo-beta-lactamase VIM-2, ... | | Authors: | Li, G.-B, Brem, J, Someya, H, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2016-07-29 | | Release date: | 2017-02-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | NMR-filtered virtual screening leads to non-metal chelating metallo-beta-lactamase inhibitors.
Chem Sci, 8, 2017
|
|
6Y9P
 
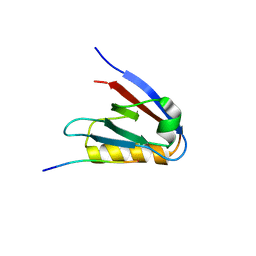 | | Crystal structure of Whirlin PDZ3_C-ter in complex with Harmonin a1 C-terminal PDZ binding motif peptide | | Descriptor: | Harmonin a1, Whirlin | | Authors: | Zhu, Y, Delhommel, F, Haouz, A, Caillet-Saguy, C, Vaney, M, Mechaly, A.E, Wolff, N. | | Deposit date: | 2020-03-10 | | Release date: | 2020-10-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.169 Å) | | Cite: | Deciphering the Unexpected Binding Capacity of the Third PDZ Domain of Whirlin to Various Cochlear Hair Cell Partners.
J.Mol.Biol., 432, 2020
|
|
8TJ9
 
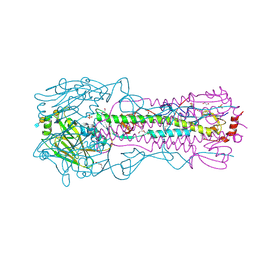 | | CRYSTAL STRUCTURE OF THE A/Michigan/15/2014(H3N2) INFLUENZA VIRUS HEMAGGLUTININ WITH HUMAN RECEPTOR ANALOG 6'-SLNLN | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wu, N.C, Zhu, X, Wilson, I.A. | | Deposit date: | 2023-07-20 | | Release date: | 2024-02-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Evolution of human H3N2 influenza virus receptor specificity has substantially expanded the receptor-binding domain site.
Cell Host Microbe, 32, 2024
|
|
6OOS
 
 | | HIV-1 Protease NL4-3 L90M Mutant in complex with darunavir | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, NL4-3 PROTEASE, SULFATE ION | | Authors: | Henes, M, Kosovrasti, K, Lockbaum, G.J, Leidner, F, Nachum, G.S, Nalivaika, E.A, Bolon, D.N.A, KurtYilmaz, N, Schiffer, C.A, Whitfield, T.W. | | Deposit date: | 2019-04-23 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular Determinants of Epistasis in HIV-1 Protease: Elucidating the Interdependence of L89V and L90M Mutations in Resistance.
Biochemistry, 58, 2019
|
|
8EZS
 
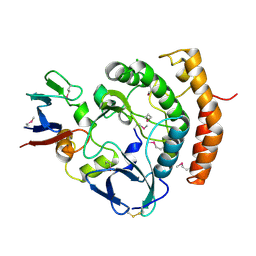 | | Crystal structure of the HipS(Lp)-HipT(Lp) complex from Legionella pneumophila, Sel-met protein | | Descriptor: | CHLORIDE ION, HipS(Lp), HipT(Lp) | | Authors: | Stogios, P.J, Skarina, T, Michalska, K, Di Leo, R, Lin, J, Ensminger, A, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-11-01 | | Release date: | 2023-09-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Crystal structure of the HipS(Lp)-HipT(Lp) complex from Legionella pneumophila, Sel-met protein
To Be Published
|
|
7S83
 
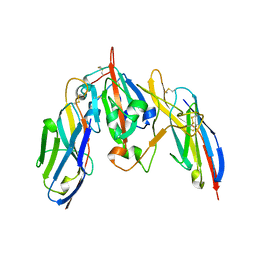 | | Crystal structure of SARS CoV-2 Spike Receptor Binding Domain in complex with shark neutralizing VNARs ShAb01 and ShAb02 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ShAb01 VNAR, ... | | Authors: | Chen, W.-H, Hajduczki, A, Dooley, H.M, Joyce, M.G. | | Deposit date: | 2021-09-17 | | Release date: | 2022-11-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Shark nanobodies with potent SARS-CoV-2 neutralizing activity and broad sarbecovirus reactivity.
Nat Commun, 14, 2023
|
|
8TJA
 
 | | CRYSTAL STRUCTURE OF THE A/Ecuador/1374/2016(H3N2) INFLUENZA VIRUS HEMAGGLUTININ WITH HUMAN RECEPTOR ANALOG 6'-SLNLN | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wu, N.C, Zhu, X, Wilson, I.A. | | Deposit date: | 2023-07-20 | | Release date: | 2024-02-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Evolution of human H3N2 influenza virus receptor specificity has substantially expanded the receptor-binding domain site.
Cell Host Microbe, 32, 2024
|
|
6YAK
 
 | | Split gene transketolase, active alpha2beta2 heterotetramer | | Descriptor: | (2S)-2-hydroxybutanedioic acid, 2-[3-[(4-azanyl-2-methyl-pyrimidin-5-yl)methyl]-4-methyl-2H-1,3-thiazol-5-yl]ethyl phosphono hydrogen phosphate, C-terminal component of the split chain transketolase, ... | | Authors: | Isupov, M.N, Littlechild, J.A, James, P. | | Deposit date: | 2020-03-12 | | Release date: | 2020-11-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | A 'Split-Gene' Transketolase From the Hyper-Thermophilic Bacterium Carboxydothermus hydrogenoformans : Structure and Biochemical Characterization.
Front Microbiol, 11, 2020
|
|
6OP7
 
 | | Structure of oxidized VIM-20 | | Descriptor: | ACETATE ION, Metallo-beta-lactamase VIM-20, ZINC ION | | Authors: | Page, R.C, Shurina, B.A, Montgomery, J.S, Orischak, M.G, Nix, J.C. | | Deposit date: | 2019-04-24 | | Release date: | 2019-10-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | A Single Salt Bridge in VIM-20 Increases Protein Stability and Antibiotic Resistance under Low-Zinc Conditions.
Mbio, 10, 2019
|
|
6Y3B
 
 | | Crystal Structure of Unlinked NS2B-NS3 Protease from Zika Virus in Complex with Inhibitor MI-2110 | | Descriptor: | 1-[(8~{R},15~{S},18~{S})-15,18-bis(4-azanylbutyl)-4,7,14,17,20-pentakis(oxidanylidene)-3,6,13,16,19-pentazabicyclo[20.3.1]hexacosa-1(25),22(26),23-trien-8-yl]guanidine, GLYCEROL, Genome polyprotein, ... | | Authors: | Huber, S, Heine, A, Steinmetzer, T. | | Deposit date: | 2020-02-18 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structure-Based Macrocyclization of Substrate Analogue NS2B-NS3 Protease Inhibitors of Zika, West Nile and Dengue viruses.
Chemmedchem, 15, 2020
|
|
7N0J
 
 | | Structure of YebY from E. coli K12 | | Descriptor: | YebY | | Authors: | Hadley, R.C, Rosenzweig, A.C. | | Deposit date: | 2021-05-25 | | Release date: | 2022-03-09 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | The copper-linked Escherichia coli AZY operon: Structure, metal binding, and a possible physiological role in copper delivery.
J.Biol.Chem., 298, 2022
|
|
8ERN
 
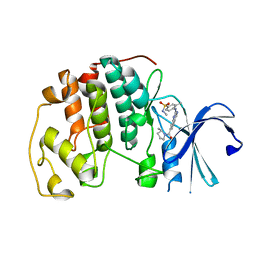 | | Cyclin-free CDK2 in complex with Cpd21 | | Descriptor: | Cyclin-dependent kinase 2, GLYCEROL, N,3-dimethyl-4-({7-[(2S)-2-methylpyrrolidine-1-carbonyl]quinazolin-2-yl}amino)benzene-1-sulfonamide | | Authors: | Murray, J.M, Oh, A. | | Deposit date: | 2022-10-12 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Cyclin-free CDK2 in complex with Cpd21
To Be Published
|
|
5LLK
 
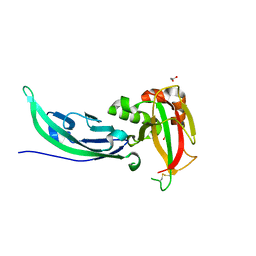 | | Crystal structure of human alpha-dystroglycan | | Descriptor: | 1,2-ETHANEDIOL, Dystroglycan | | Authors: | Covaceuszach, S, Cassetta, A, Lamba, D, Brancaccio, A, Bozzi, M, Sciandra, F, Bigotti, M.G, Konarev, P.V. | | Deposit date: | 2016-07-27 | | Release date: | 2017-07-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural flexibility of human alpha-dystroglycan.
FEBS Open Bio, 7, 2017
|
|
5LRR
 
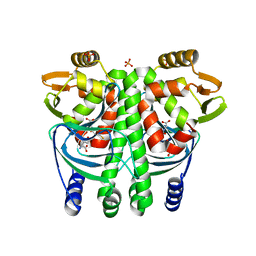 | | The Transcriptional Regulator PrfA from Listeria Monocytogenes in complex with glutathione | | Descriptor: | GLUTATHIONE, HYDROGENPHOSPHATE ION, Listeriolysin positive regulatory factor A, ... | | Authors: | Hall, M, Grundstrom, C, Begum, A, Lindberg, M, Sauer, U.H, Almqvist, F, Johansson, J, Sauer-Eriksson, A.E. | | Deposit date: | 2016-08-19 | | Release date: | 2016-12-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.171 Å) | | Cite: | Structural basis for glutathione-mediated activation of the virulence regulatory protein PrfA in Listeria.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
7PFM
 
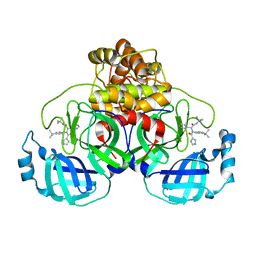 | | A SARS-CoV2 major protease non-covalent ligand structure determined to 2.0 A resolution | | Descriptor: | N-[(1R)-2-(tert-butylamino)-2-oxidanylidene-1-pyridin-3-yl-ethyl]-N-(4-tert-butylphenyl)-1H-imidazole-5-carboxamide, Replicase polyprotein 1ab | | Authors: | Moche, M, Moodie, L, Strandback, E, Nyman, T, Sandstrom, A, Akaberi, D, Lennerstrand, J. | | Deposit date: | 2021-08-11 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The SARS-CoV2 major protease (Mpro) in complex with a non-covalent inhibitory ligand at 2 A resolution
To Be Published
|
|
7NNJ
 
 | | Crystal Structure of NUDT4 (Diphosphoinositol polyphosphate phosphohydrolase 2) in complex with 4-O-Bn-1-PCP-InsP4 (AMR2105) | | Descriptor: | 1,2-ETHANEDIOL, Diphosphoinositol polyphosphate phosphohydrolase 2, FORMIC ACID, ... | | Authors: | Dubianok, Y, Arruda Bezerra, G, Raux, B, Diaz Saez, L, Riley, A.M, Potter, B.V.L, Huber, K.V.M, von Delft, F. | | Deposit date: | 2021-02-24 | | Release date: | 2022-03-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.755 Å) | | Cite: | Crystal Structure of NUDT4 (Diphosphoinositol polyphosphate phosphohydrolase 2) in complex with 4-O-Bn-1-PCP-InsP4 (AMR2105)
To Be Published
|
|
7PGD
 
 | | PAF in 50 v/v % DMSO-water solution | | Descriptor: | Pc24g00380 protein | | Authors: | Czajlik, A, Batta, G. | | Deposit date: | 2021-08-13 | | Release date: | 2022-08-24 | | Last modified: | 2023-02-15 | | Method: | SOLUTION NMR | | Cite: | DMSO-Induced Unfolding of the Antifungal Disulfide Protein PAF and Its Inactive Variant: A Combined NMR and DSC Study.
Int J Mol Sci, 24, 2023
|
|
6M8Q
 
 | | Cleavage and Polyadenylation Specificity Factor Subunit 3 (CPSF3) in complex with NVP-LTM531 | | Descriptor: | Cleavage and polyadenylation specificity factor subunit 3, N-{3,5-dichloro-2-hydroxy-4-[2-(4-methylpiperazin-1-yl)ethoxy]benzene-1-carbonyl}-L-phenylalanine, PHOSPHATE ION, ... | | Authors: | Weihofen, W.A, Salcius, M, Michaud, G. | | Deposit date: | 2018-08-22 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | CPSF3-dependent pre-mRNA processing as a druggable node in AML and Ewing's sarcoma.
Nat.Chem.Biol., 16, 2020
|
|
