6QY4
 
 | | Pink beam serial crystallography: Lysozyme, 1 us exposure, 14793 patterns merged | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C, ... | | Authors: | Lieske, J, Tolstikova, A, Meents, A. | | Deposit date: | 2019-03-08 | | Release date: | 2019-09-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | 1 kHz fixed-target serial crystallography using a multilayer monochromator and an integrating pixel detector.
Iucrj, 6, 2019
|
|
5P1Y
 
 | |
5P2C
 
 | |
5P2Q
 
 | |
8C9J
 
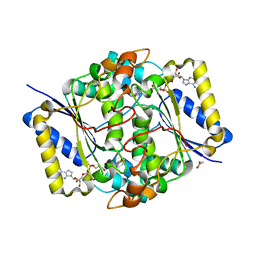 | | Crystal structure of human NQO1 by serial femtosecond crystallography | | Descriptor: | ACETATE ION, FLAVIN-ADENINE DINUCLEOTIDE, NAD(P)H dehydrogenase [quinone] 1 | | Authors: | Martin-Garcia, J.M, Grieco, A, Ruiz-Fresneda, M.A, Pacheco-Garcia, J.L, Pey, A, Botha, S, Ros, A. | | Deposit date: | 2023-01-23 | | Release date: | 2023-06-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Modular droplet injector for sample conservation providing new structural insight for the conformational heterogeneity in the disease-associated NQO1 enzyme.
Lab Chip, 23, 2023
|
|
6QYP
 
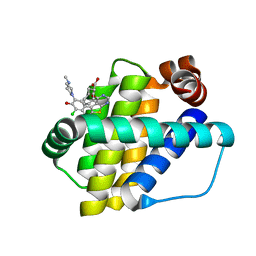 | | Structure of Mcl-1 in complex with compound 13 | | Descriptor: | (2~{R})-2-[5-[3-chloranyl-2-methyl-5-(4-methylpiperazin-1-yl)-4-oxidanyl-phenyl]-6-ethyl-thieno[2,3-d]pyrimidin-4-yl]oxy-3-phenyl-propanoic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Dokurno, P, Szlavik, Z, Ondi, L, Csekei, M, Paczal, A, Szabo, Z.B, Radics, G, Murray, J, Davidson, J, Chen, I, Davis, B, Hubbard, R.E, Pedder, C, Surgenor, A.E, Smith, J, Robertson, A, LeToumelin-Braizat, G, Cauquil, N, Zarka, M, Demarles, D, Perron-Sierra, F, Geneste, O, Kotschy, A. | | Deposit date: | 2019-03-09 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Guided Discovery of a Selective Mcl-1 Inhibitor with Cellular Activity.
J.Med.Chem., 62, 2019
|
|
6QZ5
 
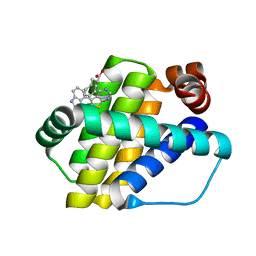 | | Structure of Mcl-1 in complex with compound 8a | | Descriptor: | (2~{R})-2-[[6-ethyl-5-(1~{H}-indol-4-yl)thieno[2,3-d]pyrimidin-4-yl]amino]-3-phenyl-propanoic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Dokurno, P, Szlavik, Z, Ondi, L, Csekei, M, Paczal, A, Szabo, Z.B, Radics, G, Murray, J, Davidson, J, Chen, I, Davis, B, Hubbard, R.E, Pedder, C, Surgenor, A.E, Smith, J, Robertson, A, LeToumelin-Braizat, G, Cauquil, N, Zarka, M, Demarles, D, Perron-Sierra, F, Geneste, O, Kotschy, A. | | Deposit date: | 2019-03-11 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Guided Discovery of a Selective Mcl-1 Inhibitor with Cellular Activity.
J.Med.Chem., 62, 2019
|
|
5P0K
 
 | |
8QK5
 
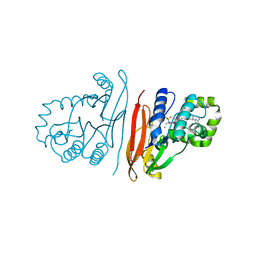 | | Structure of K. pneumoniae LpxH in complex with EBL-3647 | | Descriptor: | 5-[[3-(aminomethyl)azetidin-1-yl]methyl]-N-[4-[4-(4-cyano-6-methyl-pyrimidin-2-yl)piperazin-1-yl]sulfonylphenyl]-2-[methyl(methylsulfonyl)amino]benzamide, MANGANESE (II) ION, UDP-2,3-diacylglucosamine hydrolase | | Authors: | Sooriyaarachchi, S, Bergfors, T, Jones, T.A, Mowbray, S.L. | | Deposit date: | 2023-09-14 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Antibiotic class with potent in vivo activity targeting lipopolysaccharide synthesis in Gram-negative bacteria.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
5P34
 
 | |
5P3I
 
 | |
5P3Z
 
 | |
5P0Y
 
 | |
5P1C
 
 | |
5P1Q
 
 | |
5P26
 
 | |
6QMN
 
 | | Crystal structure of a Ribonuclease A-Onconase chimera | | Descriptor: | PHOSPHATE ION, Ribonuclease pancreatic | | Authors: | Esposito, L, Vitagliano, L, Ruggiero, A, Picone, D, Leone, S, Donnarumma, F. | | Deposit date: | 2019-02-07 | | Release date: | 2019-05-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structure, stability and aggregation propensity of a Ribonuclease A-Onconase chimera.
Int.J.Biol.Macromol., 133, 2019
|
|
5P4E
 
 | |
5P4W
 
 | |
5P5C
 
 | |
5P5R
 
 | |
5P66
 
 | |
2P5L
 
 | | Crystal structure of a dimer of N-terminal domains of AhrC in complex with an 18bp DNA operator site | | Descriptor: | Arginine repressor, DNA (5'-D(*DCP*DAP*DTP*DGP*DAP*DAP*DTP*DAP*DAP*DAP*DAP*DAP*DTP*DTP*DCP*DAP*DAP*DG)-3'), DNA (5'-D(*DCP*DTP*DTP*DGP*DAP*DAP*DTP*DTP*DTP*DTP*DTP*DAP*DTP*DTP*DCP*DAP*DTP*DG)-3'), ... | | Authors: | Garnett, J.A, Marincs, F, Baumberg, S, Stockley, P.G, Phillips, S.E.V. | | Deposit date: | 2007-03-15 | | Release date: | 2008-03-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure and function of the arginine repressor-operator complex from Bacillus subtilis.
J.Mol.Biol., 379, 2008
|
|
5P6K
 
 | |
5P2K
 
 | |
