1TJI
 
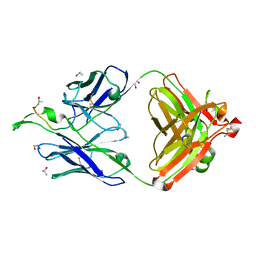 | | Crystal Structure of the broadly neutralizing anti-HIV-1 antibody 2F5 in complex with a gp41 17mer epitope | | Descriptor: | 1,2-ETHANEDIOL, Envelope Glycoprotein GP41, ISOPROPYL ALCOHOL, ... | | Authors: | Ofek, G, Tang, M, Sambor, A, Katinger, H, Mascola, J.R, Wyatt, R, Kwong, P.D. | | Deposit date: | 2004-06-04 | | Release date: | 2004-10-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and mechanistic analysis of the Anti-Human Immunodeficiency Virus type 1 antibody 2F5 in complex with its gp41 epitope
J.Virol., 78, 2004
|
|
1T06
 
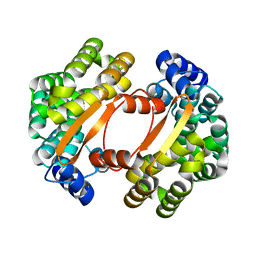 | |
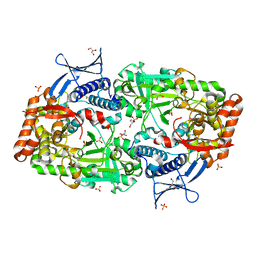
7TGJ
 
 | |
1T2M
 
 | | Solution Structure Of The Pdz Domain Of AF-6 | | Descriptor: | AF-6 protein | | Authors: | Zhou, H, Wu, J.H, Xu, Y.Q, Huang, A.D, Shi, Y.Y. | | Deposit date: | 2004-04-22 | | Release date: | 2005-02-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of AF-6 PDZ Domain and Its Interaction with the C-terminal Peptides from Neurexin and Bcr
J.Biol.Chem., 280, 2005
|
|
6JFV
 
 | | The crystal structure of 2B-2B complex from keratins 5 and 14 (C367A mutant of K14) | | Descriptor: | Keratin, type I cytoskeletal 14, type II cytoskeletal 5 | | Authors: | Kim, M.S, Lee, C.H, Coulombe, P.A, Leahy, D.J. | | Deposit date: | 2019-02-12 | | Release date: | 2020-01-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-Function Analyses of a Keratin Heterotypic Complex Identify Specific Keratin Regions Involved in Intermediate Filament Assembly.
Structure, 28, 2020
|
|
7SIH
 
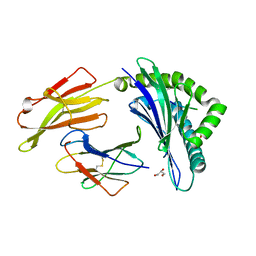 | | Crystal Structure of HLA B*3503 in complex with NPDIVIYQY, an 9-mer epitope from HIV-I | | Descriptor: | Beta-2-microglobulin, GLYCEROL, MHC class I antigen, ... | | Authors: | Gras, S, Lobos, C.A, Chatzileontiadou, D.S.M. | | Deposit date: | 2021-10-14 | | Release date: | 2022-11-23 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular insights into the HLA-B35 molecules' classification associated with HIV control.
Immunol.Cell.Biol., 102, 2024
|
|
2VIM
 
 | | X-ray structure of Fasciola hepatica thioredoxin | | Descriptor: | THIOREDOXIN | | Authors: | Line, K, Isupov, M.N, Garcia-Rodriguez, E, Maggioli, G, Parra, F, Littlechild, J.A. | | Deposit date: | 2007-12-05 | | Release date: | 2008-07-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | The Fasciola Hepatica Thioredoxin: High Resolution Structure Reveals Two Oxidation States.
Mol.Biochem.Parasitol., 161, 2008
|
|
6PAA
 
 | |
7SIG
 
 | | Crystal Structure of HLA B*3501 in complex with NPDIVIYQY, an 9-mer epitope from HIV-I | | Descriptor: | Beta-2-microglobulin, CITRATE ANION, MHC class I antigen, ... | | Authors: | Gras, S, Lobos, C.A, Chatzileontiadou, D.S.M. | | Deposit date: | 2021-10-14 | | Release date: | 2022-11-23 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.741 Å) | | Cite: | Molecular insights into the HLA-B35 molecules' classification associated with HIV control.
Immunol.Cell.Biol., 102, 2024
|
|
2I3S
 
 | | Bub3 complex with Bub1 GLEBS motif | | Descriptor: | Cell cycle arrest protein, Checkpoint serine/threonine-protein kinase | | Authors: | Larsen, N.A, Harrison, S.C. | | Deposit date: | 2006-08-20 | | Release date: | 2007-01-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural analysis of Bub3 interactions in the mitotic spindle checkpoint.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
7SJG
 
 | | Structure of Dehaloperoxidase B in Complex with Thymoquinone | | Descriptor: | (6R)-4-hydroxy-6-methyl-3-(propan-2-yl)cyclohexa-2,4-dien-1-one, 2-methyl-5-(propan-2-yl)benzene-1,4-diol, Dehaloperoxidase B, ... | | Authors: | Ghiladi, R.A, de Serrano, V.S, Malewschik, T, Yun, D. | | Deposit date: | 2021-10-17 | | Release date: | 2022-11-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Multifunctional Globin Dehaloperoxidase as a Biocatalyst in the Oxidation of Monoterpenes
To Be Published
|
|
2I79
 
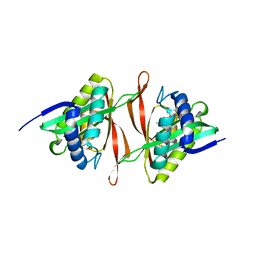 | | The crystal structure of the acetyltransferase of GNAT family from Streptococcus pneumoniae | | Descriptor: | ACETYL COENZYME *A, Acetyltransferase, GNAT family | | Authors: | Zhang, R.G, Zhou, M, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-08-30 | | Release date: | 2006-10-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of the acetyltransferase of GNAT family from Streptococcus pneumoniae
To be Published
|
|
2I8C
 
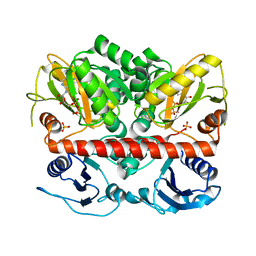 | | Allosteric inhibition of Staphylococcus aureus D-alanine:D-alanine ligase revealed by crystallographic studies | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, D-alanine-D-alanine ligase, MAGNESIUM ION, ... | | Authors: | Liu, S, Chang, J.S, Herberg, J.T, Horng, M, Tomich, P.K, Lin, A.H, Marotti, K.R. | | Deposit date: | 2006-09-01 | | Release date: | 2006-09-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Allosteric inhibition of Staphylococcus aureus D-alanine:D-alanine ligase revealed by crystallographic studies.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
6ICR
 
 | | LdCoroCC mutant- C482A | | Descriptor: | 1,2-ETHANEDIOL, Coronin-like protein | | Authors: | Karade, S.S, Srivastava, V.K, Ansari, A, Pratap, J.V. | | Deposit date: | 2018-09-06 | | Release date: | 2019-10-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Molecular and structural analysis of a mechanical transition of helices in the L. donovani coronin coiled-coil domain.
Int.J.Biol.Macromol., 143, 2020
|
|
6IDS
 
 | | Crystal structure of Vibrio cholerae MATE transporter VcmN D35N mutant | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, MATE family efflux transporter | | Authors: | Kusakizako, T, Claxton, D.P, Tanaka, Y, Maturana, A.D, Kuroda, T, Ishitani, R, Mchaourab, H.S, Nureki, O. | | Deposit date: | 2018-09-11 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structural Basis of H+-Dependent Conformational Change in a Bacterial MATE Transporter.
Structure, 27, 2019
|
|
1TI3
 
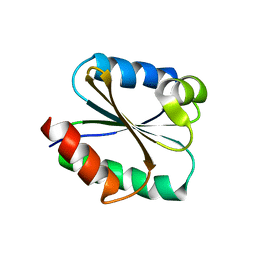 | | Solution structure of the Thioredoxin h1 from poplar, a CPPC active site variant | | Descriptor: | thioredoxin H | | Authors: | Coudevylle, N, Thureau, A, Hemmerlin, C, Gelhaye, E, Jacquot, J.P, Cung, M.T. | | Deposit date: | 2004-06-02 | | Release date: | 2004-12-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a natural CPPC active site variant, the reduced form of thioredoxin h1 from poplar.
Biochemistry, 44, 2005
|
|
6IF7
 
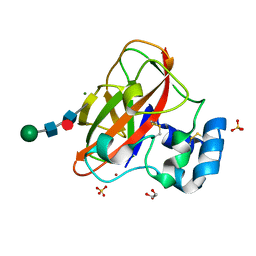 | | Crystal Structure of AA10 Lytic Polysaccharide Monooxygenase from Tectaria macrodonta | | Descriptor: | COPPER (II) ION, Chitin binding protein, GLYCEROL, ... | | Authors: | Archana, A, Yadav, S.K, Singh, P.K, Vasudev, P.G. | | Deposit date: | 2018-09-18 | | Release date: | 2019-04-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Insecticidal fern protein Tma12 is possibly a lytic polysaccharide monooxygenase.
Planta, 249, 2019
|
|
6ICD
 
 | | REGULATION OF AN ENZYME BY PHOSPHORYLATION AT THE ACTIVE SITE | | Descriptor: | ISOCITRATE DEHYDROGENASE | | Authors: | Hurley, J.H, Dean, A.M, Sohl, J.L, Koshlandjunior, D.E, Stroud, R.M. | | Deposit date: | 1990-05-30 | | Release date: | 1991-10-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Regulation of an enzyme by phosphorylation at the active site.
Science, 249, 1990
|
|
2VD2
 
 | |
3EI1
 
 | |
2I52
 
 | | Crystal structure of protein PTO0218 from Picrophilus torridus, Pfam DUF372 | | Descriptor: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Ramagopal, U.A, Gilmore, J, Toro, R, Bain, K.T, McKenzie, C, Reyes, C, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-08-23 | | Release date: | 2006-09-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structure of hypothetical protein PTO0218 from Picrophilus torridus
To be Published
|
|
1SZA
 
 | |
3EKU
 
 | | Crystal Structure of Monomeric Actin bound to Cytochalasin D | | Descriptor: | (3S,3aR,4S,6S,6aR,7E,10S,12R,13E,15R,15aR)-3-benzyl-6,12-dihydroxy-4,10,12-trimethyl-5-methylidene-1,11-dioxo-2,3,3a,4,5,6,6a,9,10,11,12,15-dodecahydro-1H-cycloundeca[d]isoindol-15-yl acetate, ADENOSINE-5'-TRIPHOSPHATE, Actin-5C, ... | | Authors: | Nair, U.B, Joel, P.B, Wan, Q, Lowey, S, Rould, M.A, Trybus, K.M. | | Deposit date: | 2008-09-19 | | Release date: | 2008-10-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of monomeric actin bound to cytochalasin D.
J.Mol.Biol., 384, 2008
|
|
1T10
 
 | |
8IJX
 
 | | Cryo-EM structure of the gastric proton pump with bound DQ-18 | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-[4-[(5-chloranyl-2-phenylmethoxy-phenyl)methoxy]phenyl]-N-methyl-methanamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Abe, K, Yokoshima, S, Yoshimori, A. | | Deposit date: | 2023-02-28 | | Release date: | 2023-08-30 | | Last modified: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (2.08 Å) | | Cite: | Deep learning driven de novo drug design based on gastric proton pump structures.
Commun Biol, 6, 2023
|
|
