8G8D
 
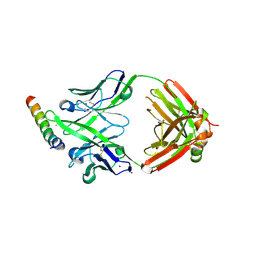 | | Crystal structure of DH1346 Fab in complex with HIV proximal MPER peptide | | Descriptor: | DH1346 heavy chain, DH1346 light chain, FLUORIDE ION, ... | | Authors: | Niyongabo, A, Janus, B.M, Ofek, G. | | Deposit date: | 2023-02-17 | | Release date: | 2024-05-22 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Vaccine induction of heterologous HIV-1-neutralizing antibody B cell lineages in humans.
Cell, 187, 2024
|
|
5NW3
 
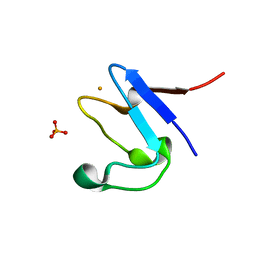 | | The cryofrozen atomic resolution X-ray crystal structure of perdeuterated Pyrococcus furiosus Rubredoxin (100K, 0.59A resolution) | | Descriptor: | FE (III) ION, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Cuypers, M.G, Mason, S.A, Mossou, E, Haertlein, M, Forsyth, V.T. | | Deposit date: | 2017-05-04 | | Release date: | 2017-06-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (0.59 Å) | | Cite: | The cryofrozen atomic resolution X-ray crystal structure of perdeuterated Pyrococcus furiosus Rubredoxin (100K, 0.59A resolution)
To Be Published
|
|
5MTK
 
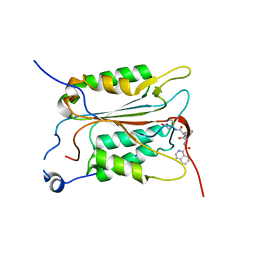 | | Crystal structure of human Caspase-1 with (3S,6S,10aS)-N-((2S,3S)-2-hydroxy-5-oxotetrahydrofuran-3-yl)-6-(isoquinoline-1-carboxamido)-5-oxodecahydropyrrolo[1,2-a]azocine-3-carboxamide (PGE-3935199) | | Descriptor: | (3~{S})-3-[[(3~{S},6~{S},10~{a}~{S})-6-(isoquinolin-1-ylcarbonylamino)-5-oxidanylidene-2,3,6,7,8,9,10,10~{a}-octahydro-1~{H}-pyrrolo[1,2-a]azocin-3-yl]carbonylamino]-4-oxidanyl-butanoic acid, Caspase-1 | | Authors: | Brethon, A, Chantalat, L, Christin, O, Clary, L, Fournier, J.F, Gastreich, M, Harris, C, Pascau, J, Isabet, T, Rodeschin, V, Thoreau, E, Roche, D. | | Deposit date: | 2017-01-09 | | Release date: | 2018-02-28 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Playing against the odds: scaffold hopping from 3D-fragments
To Be Published
|
|
3S90
 
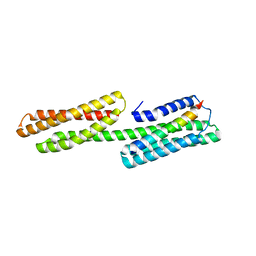 | | Human vinculin head domain Vh1 (residues 1-252) in complex with murine talin (VBS33; residues 1512-1546) | | Descriptor: | Talin-1, Vinculin | | Authors: | Yogesha, S.D, Sharff, A, Bricogne, G, Izard, T. | | Deposit date: | 2011-05-31 | | Release date: | 2011-06-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Intermolecular versus intramolecular interactions of the vinculin binding site 33 of talin.
Protein Sci., 20, 2011
|
|
3OOH
 
 | | Crystal structure of E. Coli purine nucleoside phosphorylase with PO4 | | Descriptor: | PHOSPHATE ION, Purine nucleoside phosphorylase deoD-type | | Authors: | Mikleusevic, G, Stefanic, Z, Narzyk, M, Wielgus-Kutrowska, B, Bzowska, A, Luic, M. | | Deposit date: | 2010-08-31 | | Release date: | 2011-07-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Validation of the catalytic mechanism of Escherichia coli purine nucleoside phosphorylase by structural and kinetic studies.
Biochimie, 93, 2011
|
|
8G8A
 
 | | Crystal structure of DH1317.8 Fab in complex with HIV proximal MPER peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DH1317.8 heavy chain, DH1317.8 light chain, ... | | Authors: | Janus, B.M, Astavans, A, Ofek, G. | | Deposit date: | 2023-02-17 | | Release date: | 2024-05-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Vaccine induction of heterologous HIV-1-neutralizing antibody B cell lineages in humans.
Cell, 187, 2024
|
|
7A06
 
 | | Structure of human CKa1 in complex with compound o | | Descriptor: | 1,2-ETHANEDIOL, 1-[(4-phenylphenyl)methyl]-4-pyrrolidin-1-yl-pyridine, Choline kinase alpha | | Authors: | Serran-Aguilera, L, Mariotto, E, Rubbini, G, Castro Navas, F.C, Marco, C, Carrasco-Jimenez, M.P, Ballarotto, M, Macchiarulo, A, Hurtado-Guerrero, R, Viola, G, Lopez-Cara, L.C. | | Deposit date: | 2020-08-06 | | Release date: | 2020-09-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Synthesis, biological evaluation, in silico modeling and crystallization of novel small monocationic molecules with potent antiproliferative activity by dual mechanism.
Eur.J.Med.Chem., 207, 2020
|
|
8DYN
 
 | |
6SDP
 
 | |
5MBT
 
 | | CeuE (H227L, Y288F variant) a periplasmic protein from Campylobacter jejuni | | Descriptor: | Enterochelin uptake periplasmic binding protein | | Authors: | Wilde, E.J, Blagova, E.V, Hughes, A, Raines, D.J, Moroz, O.V, Turkenburg, J.P, Duhme-Klair, A.-K, Wilson, K.S. | | Deposit date: | 2016-11-08 | | Release date: | 2017-04-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Interactions of the periplasmic binding protein CeuE with Fe(III) n-LICAM(4-) siderophore analogues of varied linker length.
Sci Rep, 7, 2017
|
|
5MUP
 
 | | Structure of deformed wing virus, a honeybee pathogen | | Descriptor: | URIDINE-5'-MONOPHOSPHATE, VP1, VP2, ... | | Authors: | Skubnik, K, Novacek, J, Fuzik, T, Pridal, A, Paxton, R, Plevka, P. | | Deposit date: | 2017-01-13 | | Release date: | 2017-04-05 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of deformed wing virus, a major honey bee pathogen.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7XMP
 
 | |
8DQG
 
 | | Crystal structure of pyrrolysyl-tRNA synthetase from Methanomethylophilus alvus engineered for acridone amino acid (RS1) bound to AMPPNP and acridone | | Descriptor: | (2~{S})-2-azanyl-3-(9-oxidanylidene-10~{H}-acridin-2-yl)propanoic acid, AA_TRNA_LIGASE_II domain-containing protein, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Gottfried-Lee, I, Karplus, P.A, Mehl, R.A, Cooley, R.B. | | Deposit date: | 2022-07-19 | | Release date: | 2022-12-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structures of Methanomethylophilus alvus Pyrrolysine tRNA-Synthetases Support the Need for De Novo Selections When Altering the Substrate Specificity.
Acs Chem.Biol., 17, 2022
|
|
2WK5
 
 | | Structural features of native human thymidine phosphorylase and in complex with 5-iodouracil | | Descriptor: | GLYCEROL, THYMIDINE PHOSPHORYLASE | | Authors: | Mitsiki, E, Papageorgiou, A.C, Iyer, S, Thiyagarajan, N, Prior, S.H, Sleep, D, Finnis, C, Acharya, K.R. | | Deposit date: | 2009-06-05 | | Release date: | 2009-07-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Structures of Native Human Thymidine Phosphorylase and in Complex with 5-Iodouracil.
Biochem.Biophys.Res.Commun., 386, 2009
|
|
8Q5P
 
 | | Structure of the lysine methyltransferase SETD2 in complex with a peptide derived from human tyrosine kinase ACK1 | | Descriptor: | Activated CDC42 kinase 1, Histone-lysine N-methyltransferase SETD2, S-ADENOSYLMETHIONINE, ... | | Authors: | Mechaly, A, Le Coadou, L, Dupret, J.M, Haouz, A, Rodrigues Lima, F. | | Deposit date: | 2023-08-09 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.814 Å) | | Cite: | Structural and enzymatic evidence for the methylation of the ACK1 tyrosine kinase by the histone lysine methyltransferase SETD2.
Biochem.Biophys.Res.Commun., 695, 2024
|
|
7FZS
 
 | | Crystal Structure of human FABP4 in complex with 6-chloro-4-[(2-chlorophenyl)methyl]-2-methylquinoline-3-carboxylic acid | | Descriptor: | 6-chloro-4-[(2-chlorophenyl)methyl]-2-methylquinoline-3-carboxylic acid, Fatty acid-binding protein, adipocyte, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Conte, A, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Crystal Structure of a human FABP4 complex
To be published
|
|
5MV5
 
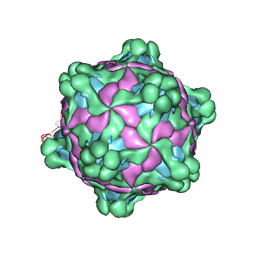 | | Structure of deformed wing virus, a honeybee pathogen | | Descriptor: | URIDINE-5'-MONOPHOSPHATE, VP1, VP2, ... | | Authors: | Skubnik, K, Novacek, J, Fuzik, T, Pridal, A, Paxton, R, Plevka, P. | | Deposit date: | 2017-01-15 | | Release date: | 2017-04-05 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of deformed wing virus, a major honey bee pathogen.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6SAW
 
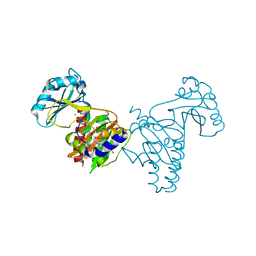 | |
8QKA
 
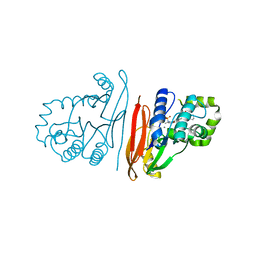 | | Structure of K. pneumoniae LpxH in complex with JEDI-852 | | Descriptor: | 2-[methyl(methylsulfonyl)amino]-~{N}-(4-piperidin-1-ylsulfonylphenyl)benzamide, MANGANESE (II) ION, UDP-2,3-diacylglucosamine hydrolase | | Authors: | Sooriyaarachchi, S, Bergfors, T, Jones, T.A, Mowbray, S.L. | | Deposit date: | 2023-09-14 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Antibiotic class with potent in vivo activity targeting lipopolysaccharide synthesis in Gram-negative bacteria.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8E3U
 
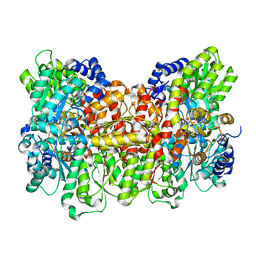 | |
6BSF
 
 | | Human GR (418-507) in complex with monomeric DNA binding site | | Descriptor: | DNA (5'-D(P*AP*AP*GP*CP*TP*AP*GP*TP*AP*CP*AP*TP*TP*TP*GP*C)-3'), DNA (5'-D(P*TP*GP*CP*AP*AP*AP*TP*GP*TP*AP*CP*TP*AP*GP*CP*T)-3'), Glucocorticoid receptor, ... | | Authors: | Pufall, M.A. | | Deposit date: | 2017-12-02 | | Release date: | 2018-12-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | General and sequence-specific roles for DNA in glucocorticoid receptor DNA-binding stoichiometry
To Be Published
|
|
5O8I
 
 | |
5CUC
 
 | | Crystal structure of the bromodomain of bromodomain adjacent to zinc finger domain protein 2B (BAZ2B) in complex with N-Acetyl-2-phenylethylamine (SGC - Diamond I04-1 fragment screening) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain adjacent to zinc finger domain protein 2B, N-(2-phenylethyl)acetamide | | Authors: | Bradley, A, Pearce, N, Krojer, T, Ng, J, Talon, R, Vollmar, M, Jose, B, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-07-24 | | Release date: | 2015-09-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of the second bromodomain of bromodomain adjancent to zinc finger domain protein 2B (BAZ2B) in complex with N-Acetyl-2-phenylethylamine (SGC - Diamond I04-1 fragment screening)
To be published
|
|
6XJ6
 
 | |
6P4X
 
 | |
