6O0Y
 
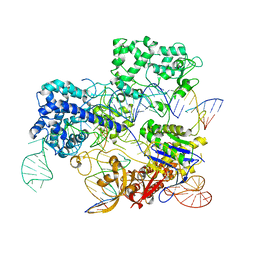 | | Conformational states of Cas9-sgRNA-DNA ternary complex in the presence of magnesium | | Descriptor: | 3' product of target strand DNA, 5' product of target strand DNA, CRISPR-associated endonuclease Cas9/Csn1, ... | | Authors: | Zhu, X, Clarke, R, Puppala, A.K, Chittori, S, Merk, A, Merrill, B.J, Simonovic, M, Subramaniam, S. | | Deposit date: | 2019-02-17 | | Release date: | 2019-07-10 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Cryo-EM structures reveal coordinated domain motions that govern DNA cleavage by Cas9.
Nat.Struct.Mol.Biol., 26, 2019
|
|
3B5M
 
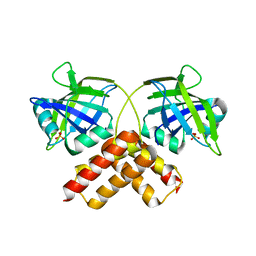 | | Crystal structure of conserved uncharacterized protein from Rhodopirellula baltica | | Descriptor: | SULFATE ION, Uncharacterized protein | | Authors: | Patskovsky, Y, Bonanno, J.B, Sridhar, V, Rutter, M, Powell, A, Maletic, M, Rodgers, R, Wasserman, S, Smith, D, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-10-26 | | Release date: | 2007-11-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Crystal Structure of Conserved Uncharacterized Protein from Rhodopirellula baltica.
To be Published
|
|
7ML6
 
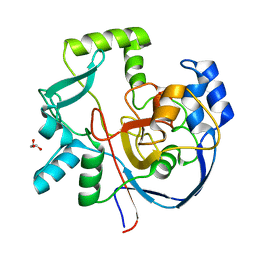 | | Structure of CalU17 from the Calicheamicin Biosynthesis Pathway of Micromonospora echinospora | | Descriptor: | CalU17, GLYCEROL | | Authors: | Kosgei, A.J, Miller, M.D, Xu, W, Van Lanen, S.G, Thorson, J.S, Phillips Jr, G.N. | | Deposit date: | 2021-04-27 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of DynF from the dynemicin-biosynthesis pathway of Micromonospora chersina.
Acta Crystallogr.,Sect.F, 78, 2022
|
|
6VGW
 
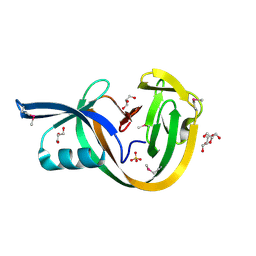 | | Crystal structure of VidaL intein (selenomethionine variant) | | Descriptor: | GLYCEROL, SULFATE ION, VidaL | | Authors: | Burton, A.J, Haugbro, M, Parisi, E, Muir, T.W. | | Deposit date: | 2020-01-09 | | Release date: | 2020-05-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Live-cell protein engineering with an ultra-short split intein.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6VH1
 
 | | 2.30 A resolution structure of MERS 3CL protease in complex with inhibitor 6h | | Descriptor: | N~2~-{[(4,4-difluorocyclohexyl)oxy]carbonyl}-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide, Orf1a protein | | Authors: | Lovell, S, Battaile, K.P, Kashipathy, M.M, Rathnayake, A.D, Zheng, J, Kim, Y, Nguyen, H.N, Chang, K.O, Groutas, W.C. | | Deposit date: | 2020-01-09 | | Release date: | 2020-08-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | 3C-like protease inhibitors block coronavirus replication in vitro and improve survival in MERS-CoV-infected mice.
Sci Transl Med, 12, 2020
|
|
5MPZ
 
 | |
1BD7
 
 | | CIRCULARLY PERMUTED BB2-CRYSTALLIN | | Descriptor: | CIRCULARLY PERMUTED BB2-CRYSTALLIN | | Authors: | Wright, G, Basak, A.K, Mayr, E.-M, Glockshuber, R, Slingsby, C. | | Deposit date: | 1998-05-12 | | Release date: | 1998-10-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Circular permutation of betaB2-crystallin changes the hierarchy of domain assembly.
Protein Sci., 7, 1998
|
|
3B73
 
 | |
8WSA
 
 | | Cryo-EM Structure of Mouse TLR4/MD-2/DLAM5 Complex | | Descriptor: | (3R)-3-(dodecanoyloxy)tetradecanoic acid, (3R)-3-(tetradecanoyloxy)tetradecanoic acid, (3~{S})-3-decanoyloxytetradecanoic acid, ... | | Authors: | Fu, Y, Kim, H, Zamyatina, A, Kim, H.M. | | Deposit date: | 2023-10-17 | | Release date: | 2024-10-23 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insight into TLR4/MD-2 activation by synthetic LPS mimetics with distinct binding modes.
Nat Commun, 16, 2025
|
|
7R3Y
 
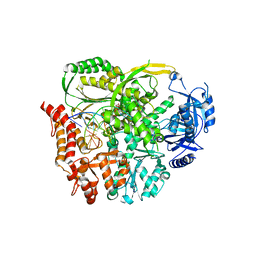 | | The crystal structure of the V426L variant of Pol2CORE in complex with DNA and an incoming nucleotide | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, CALCIUM ION, DNA Primer, ... | | Authors: | Barbari, S.R, Beach, A.K, Markgren, J.G, Parkash, V, Johansson, E, Shcherbakova, P.V. | | Deposit date: | 2022-02-08 | | Release date: | 2022-07-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Enhanced polymerase activity permits efficient synthesis by cancer-associated DNA polymerase ε variants at low dNTP levels.
Nucleic Acids Res., 50, 2022
|
|
5MPE
 
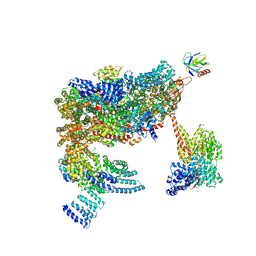 | | 26S proteasome in presence of ATP (s2) | | Descriptor: | 26S proteasome complex subunit SEM1, 26S proteasome regulatory subunit RPN1, 26S proteasome regulatory subunit RPN10, ... | | Authors: | Wehmer, M, Rudack, T, Beck, F, Aufderheide, A, Pfeifer, G, Plitzko, J.M, Foerster, F, Schulten, K, Baumeister, W, Sakata, E. | | Deposit date: | 2016-12-16 | | Release date: | 2017-03-08 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural insights into the functional cycle of the ATPase module of the 26S proteasome.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5MQG
 
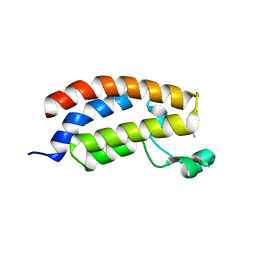 | |
6NXA
 
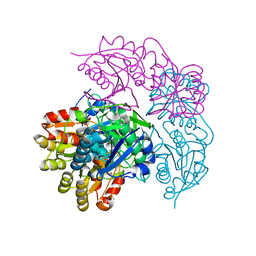 | | ECAII(D90T,K162T) MUTANT AT PH 7 | | Descriptor: | ACETIC ACID, GLYCEROL, L-asparaginase 2 | | Authors: | Lubkowski, J, Wlodawer, A. | | Deposit date: | 2019-02-08 | | Release date: | 2019-08-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Opportunistic complexes of E. coli L-asparaginases with citrate anions.
Sci Rep, 9, 2019
|
|
1FOZ
 
 | | STRUCTURE OF CYCLIC PEPTIDE INHIBITORS OF MAMMALIAN RIBONUCLEOTIDE REDUCTASE | | Descriptor: | SYNTHETIC CYCLIC PEPTIDE | | Authors: | Pellegrini, M, Liehr, S, Fisher, A.L, Cooperman, B.S, Mierke, D.F. | | Deposit date: | 2000-08-29 | | Release date: | 2000-11-22 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Structure-based optimization of peptide inhibitors of mammalian ribonucleotide reductase.
Biochemistry, 39, 2000
|
|
6OHW
 
 | | Structural basis for human coronavirus attachment to sialic acid receptors. Apo-HCoV-OC43 S | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike surface glycoprotein, ... | | Authors: | Tortorici, M.A, Walls, A.C, Lang, Y, Wang, C, Li, Z, Koerhuis, D, Boons, G.J, Bosch, B.J, Rey, F.A, de Groot, R, Veesler, D. | | Deposit date: | 2019-04-07 | | Release date: | 2019-06-05 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for human coronavirus attachment to sialic acid receptors.
Nat.Struct.Mol.Biol., 26, 2019
|
|
7NEY
 
 | | Structure of T. atroviride Fdc wild-type (TaFdc) in complex with prFMN | | Descriptor: | 1-deoxy-5-O-phosphono-1-(3,3,4,5-tetramethyl-9,11-dioxo-2,3,8,9,10,11-hexahydro-7H-quinolino[1,8-fg]pteridin-12-ium-7-y l)-D-ribitol, Ferulic acid decarboxylase 1, MANGANESE (II) ION, ... | | Authors: | Saaret, A, Leys, D. | | Deposit date: | 2021-02-05 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Directed evolution of prenylated FMN-dependent Fdc supports efficient in vivo isobutene production.
Nat Commun, 12, 2021
|
|
8Q8G
 
 | | Crystal structure of HsRNMT complexed with inhibitor DDD1870799 | | Descriptor: | 2-(4-fluorophenyl)-1-[(3~{S})-3-(6-oxidanyl-1~{H}-pyrazolo[3,4-b]pyridin-3-yl)piperidin-1-yl]ethanone, GLYCEROL, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Petit, A.P, Fyfe, P.K. | | Deposit date: | 2023-08-18 | | Release date: | 2024-08-28 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Characterisation of RNA guanine-7 methyltransferase (RNMT) using a small molecule approach.
Biochem.J., 482, 2025
|
|
6H1O
 
 | | Structure of the BM3 heme domain in complex with voriconazole | | Descriptor: | 1,2-ETHANEDIOL, Bifunctional cytochrome P450/NADPH--P450 reductase, GLYCEROL, ... | | Authors: | Jeffreys, L.N, Munro, A.W.M, Leys, D. | | Deposit date: | 2018-07-12 | | Release date: | 2019-02-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.734 Å) | | Cite: | Novel insights into P450 BM3 interactions with FDA-approved antifungal azole drugs.
Sci Rep, 9, 2019
|
|
8HCM
 
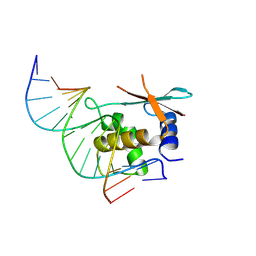 | | zebrafish IRF-11 DBD complex with DNA | | Descriptor: | DNA (5'-D(P*GP*CP*TP*TP*TP*CP*AP*CP*TP*TP*TP*CP*TP*A)-3'), DNA (5'-D(P*TP*AP*GP*AP*AP*AP*GP*TP*GP*AP*AP*AP*GP*C)-3'), Interferon regulatory factor | | Authors: | Wang, Z.X, Zhang, Y.A, Ouyang, S.Y. | | Deposit date: | 2022-11-02 | | Release date: | 2024-05-15 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal Structures of DNA-bound Fish IRF10 and IRF11 Reveal the Determinants of IFN Regulation.
J Immunol., 213, 2024
|
|
1AX3
 
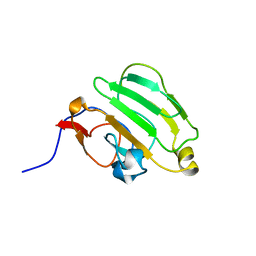 | | SOLUTION NMR STRUCTURE OF B. SUBTILIS IIAGLC, 16 STRUCTURES | | Descriptor: | GLUCOSE PERMEASE IIA DOMAIN | | Authors: | Chen, Y, Case, D.A, Reizer, J, Saier Junior, M.H, Wright, P.E. | | Deposit date: | 1997-10-25 | | Release date: | 1998-06-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | High-resolution solution structure of Bacillus subtilis IIAglc.
Proteins, 31, 1998
|
|
7NF0
 
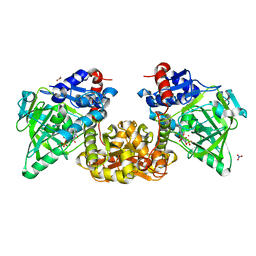 | |
4NOT
 
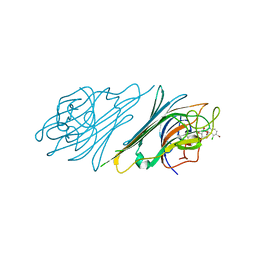 | | Crystal structure of Dioclea sclerocarpa lectin complexed with X-man | | Descriptor: | 5-bromo-4-chloro-1H-indol-3-yl alpha-D-mannopyranoside, CALCIUM ION, Lectin alpha chain, ... | | Authors: | Barroso-Neto, I.L, Teixeira, C.S, Correia, J.L.A, Santiago, M.Q, Pinto-Junior, V.R, Osterne, V.J.S, Plinio, D, Rocha, B.A.M, Cavada, B.S. | | Deposit date: | 2013-11-20 | | Release date: | 2015-01-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of Dioclea sclerocarpa lectin complexed with X-man
To be Published
|
|
3FGR
 
 | | Two chain form of the 66.3 kDa protein at 1.8 Angstroem | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Lakomek, K, Dickmanns, A, Ficner, R. | | Deposit date: | 2008-12-08 | | Release date: | 2009-09-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Initial insight into the function of the lysosomal 66.3 kDa protein from mouse by means of X-ray crystallography
Bmc Struct.Biol., 9, 2009
|
|
7DVF
 
 | | Crystal structure of the computationally designed reDPBB_sym2 protein | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, reDPBB_sym2 protein | | Authors: | Yagi, S, Tagami, S, Padhi, A.K, Zhang, K.Y.J. | | Deposit date: | 2021-01-13 | | Release date: | 2021-10-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.209 Å) | | Cite: | Seven Amino Acid Types Suffice to Create the Core Fold of RNA Polymerase.
J.Am.Chem.Soc., 143, 2021
|
|
5M8B
 
 | |
