5U4V
 
 | |
5GKF
 
 | | Structure of EndoMS-dsDNA1' complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA (5'-D(*CP*GP*CP*TP*AP*CP*AP*TP*GP*TP*CP*GP*TP*CP*C)-3'), DNA (5'-D(*GP*GP*AP*CP*GP*AP*CP*TP*TP*GP*TP*AP*GP*CP*G)-3'), ... | | Authors: | Nakae, S, Hijikata, A, Tsuji, T, Yonezawa, K, Kouyama, K, Mayanagi, K, Ishino, S, Ishino, Y, Shirai, T. | | Deposit date: | 2016-07-04 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the EndoMS-DNA Complex as Mismatch Restriction Endonuclease
Structure, 24, 2016
|
|
3MLF
 
 | |
8DTS
 
 | |
3FLS
 
 | |
3FLZ
 
 | |
7N1Z
 
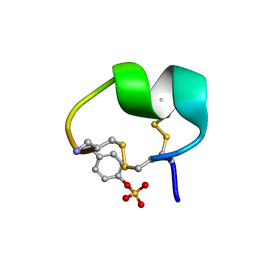 | | NMR structure of native PnIA | | Descriptor: | Alpha-conotoxin PnIA | | Authors: | Conibear, A.C, Rosengren, K.J, Lee, H.S. | | Deposit date: | 2021-05-28 | | Release date: | 2021-11-17 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Posttranslational modifications of alpha-conotoxins: sulfotyrosine and C-terminal amidation stabilise structures and increase acetylcholine receptor binding.
Rsc Med Chem, 12, 2021
|
|
7XQ7
 
 | | The complex structure of WT-Mpro | | Descriptor: | 3C-like proteinase nsp5, SODIUM ION | | Authors: | Sahoo, P, Lenka, D.R, Kumar, A. | | Deposit date: | 2022-05-06 | | Release date: | 2023-03-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Detailed Insights into the Inhibitory Mechanism of New Ebselen Derivatives against Main Protease (M pro ) of Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2).
Acs Pharmacol Transl Sci, 6, 2023
|
|
5NIU
 
 | | Structure of human Programmed cell death 1 ligand 1 (PD-L1) with low molecular mass inhibitor | | Descriptor: | (2~{R})-2-[[2-[(3-cyanophenyl)methoxy]-4-[[3-(2,3-dihydro-1,4-benzodioxin-6-yl)-2-methyl-phenyl]methoxy]-5-methyl-phenyl]methylamino]-3-oxidanyl-propanoic acid, 1,2-ETHANEDIOL, Programmed cell death 1 ligand 1 | | Authors: | Zak, K.M, Grudnik, P, Skalniak, L, Dubin, G, Holak, T.A. | | Deposit date: | 2017-03-27 | | Release date: | 2017-12-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Small-molecule inhibitors of PD-1/PD-L1 immune checkpoint alleviate the PD-L1-induced exhaustion of T-cells.
Oncotarget, 8, 2017
|
|
4YFN
 
 | | Escherichia coli RNA polymerase in complex with squaramide compound 14 (N-[3,4-dioxo-2-(4-{[4-(trifluoromethyl)benzyl]amino}piperidin-1-yl)cyclobut-1-en-1-yl]-3,5-dimethyl-1,2-oxazole-4-sulfonamide) | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Molodtsov, V, Fleming, P.R, Eyermann, C.J, Ferguson, A.D, Foulk, M.A, McKinney, D.C, Masse, C.E, Buurman, E.T, Murakami, K.S. | | Deposit date: | 2015-02-25 | | Release date: | 2015-03-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.817 Å) | | Cite: | X-ray Crystal Structures of Escherichia coli RNA Polymerase with Switch Region Binding Inhibitors Enable Rational Design of Squaramides with an Improved Fraction Unbound to Human Plasma Protein.
J.Med.Chem., 58, 2015
|
|
8U9L
 
 | | Crystal Structure of RelA-cRel chimera complex with DNA | | Descriptor: | DNA (5'-D(P*GP*AP*AP*TP*CP*GP*GP*AP*AP*AP*TP*TP*CP*CP*CP*AP*TP*CP*AP*A)-3'), DNA (5'-D(P*TP*TP*GP*AP*TP*GP*GP*GP*AP*AP*TP*TP*TP*CP*CP*GP*AP*TP*TP*C)-3'), Transcription factor p65,Proto-oncogene c-Rel chimera | | Authors: | Chang, A, Wu, Y, Li, S.X, Smale, S, Chen, L. | | Deposit date: | 2023-09-19 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (3.091 Å) | | Cite: | Crystal Structure of RelA-cRel chimera complex with DNA
To Be Published
|
|
5NEY
 
 | | Discovery, crystal structures and atomic force microscopy study of thioether ligated D,L-cyclic antimicrobial peptides against multidrug resistant Pseudomonas aeruginosa | | Descriptor: | 3,7-anhydro-2,8-dideoxy-L-glycero-D-gluco-octonic acid, CALCIUM ION, CYS-TRD-TRP-LYD-LYS-LYD-LYS-LYD-TRP-TRD-CYS-ALA, ... | | Authors: | Reymond, J.-L, Darbre, T, Stocker, A, Hong, W, van Delden, C, Koehler, T, Luscher, A, Visini, R, Fu, Y, Di Bonaventura, I, He, R. | | Deposit date: | 2017-03-13 | | Release date: | 2017-09-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Design, crystal structure and atomic force microscopy study of thioether ligated d,l-cyclic antimicrobial peptides against multidrug resistant Pseudomonas aeruginosa.
Chem Sci, 8, 2017
|
|
5A7U
 
 | | Single-particle cryo-EM of co-translational folded adr1 domain inside the E. coli ribosome exit tunnel. | | Descriptor: | REGULATORY PROTEIN ADR1, ZINC ION | | Authors: | Nilsson, O.B, Hedman, R, Marino, J, Wickles, S, Bischoff, L, Johansson, M, Muller-Lucks, A, Trovato, F, Puglisi, J.D, O'Brien, E, Beckmann, R, von Heijne, G. | | Deposit date: | 2015-07-10 | | Release date: | 2015-09-16 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Cotranslational Protein Folding Inside the Ribosome Exit Tunnel.
Cell Rep., 12, 2015
|
|
5M5X
 
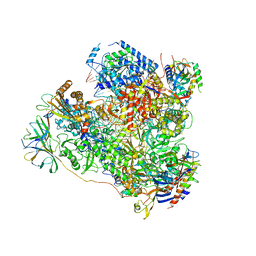 | | RNA Polymerase I elongation complex 1 | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | Authors: | Tafur, L, Sadian, Y, Hoffmann, N.A, Jakobi, A.J, Wetzel, R, Hagen, W.J.H, Sachse, C, Muller, C.W. | | Deposit date: | 2016-10-23 | | Release date: | 2016-12-21 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Molecular Structures of Transcribing RNA Polymerase I.
Mol. Cell, 64, 2016
|
|
6FJV
 
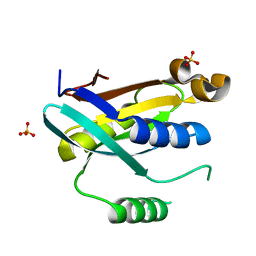 | | Rpn11 homolog from Caldiarchaeum Subterraneum, truncated | | Descriptor: | 26S proteasome regulatory subunit N11-like protein, SULFATE ION | | Authors: | Fuchs, A.C.D, Albrecht, R, Martin, J, Hartmann, M.D. | | Deposit date: | 2018-01-23 | | Release date: | 2018-07-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Rpn11-mediated ubiquitin processing in an ancestral archaeal ubiquitination system.
Nat Commun, 9, 2018
|
|
8GHZ
 
 | | Cryo-EM structure of fish immunogloblin M-Fc | | Descriptor: | Teleost immunoglobulin M protein, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Lyu, M, Stadtmueller, B.M, Malyutin, A.G. | | Deposit date: | 2023-03-13 | | Release date: | 2023-11-08 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.78 Å) | | Cite: | The structure of the teleost Immunoglobulin M core provides insights on polymeric antibody evolution, assembly, and function.
Nat Commun, 14, 2023
|
|
7XQ6
 
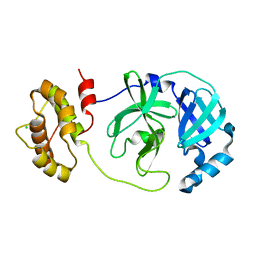 | | The complex structure of mutant Mpro with inhibitor | | Descriptor: | 3C-like proteinase nsp5, CHLORIDE ION | | Authors: | Sahoo, P, Lenka, D.R, Kumar, A. | | Deposit date: | 2022-05-06 | | Release date: | 2023-03-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Detailed Insights into the Inhibitory Mechanism of New Ebselen Derivatives against Main Protease (M pro ) of Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2).
Acs Pharmacol Transl Sci, 6, 2023
|
|
7U31
 
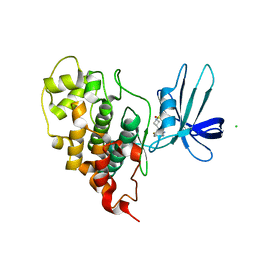 | | Crystal structure of human GSK3B in complex with G5 | | Descriptor: | 5-(4-fluorophenyl)-4-[1-(methanesulfonyl)azetidin-3-yl]pyrimidin-2-amine, CHLORIDE ION, Glycogen synthase kinase-3 beta | | Authors: | Tripathi, S.K, Balboni, B, Giabbai, B, Storici, P, Girotto, S, Cavalli, A. | | Deposit date: | 2022-02-25 | | Release date: | 2022-04-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Identification of Novel GSK-3 beta Hits Using Competitive Biophysical Assays.
Int J Mol Sci, 23, 2022
|
|
6FIE
 
 | | Crystallographic structure of calcium loaded Calbindin-D28K. | | Descriptor: | CALCIUM ION, Calbindin, THIOCYANATE ION | | Authors: | Noble, J.W, Almalki, R, Roe, S.M, Wagner, A, Dumanc, R, Atack, J.R. | | Deposit date: | 2018-01-18 | | Release date: | 2018-10-10 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | The X-ray structure of human calbindin-D28K: an improved model.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
6SYJ
 
 | |
6YJU
 
 | | Crystal structure of MGAT5 (alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase V) luminal domain with a Lys329-Ile345 loop truncation, in complex with UDP and biantennary pentasaccharide M592 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]alpha-D-mannopyranose, ... | | Authors: | Wu, L, Darby, J.F, Gilio, A.K, Davies, G.J. | | Deposit date: | 2020-04-04 | | Release date: | 2020-08-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Substrate Engagement and Catalytic Mechanisms of N-Acetylglucosaminyltransferase V
Acs Catalysis, 2020
|
|
7U36
 
 | | Crystal structure of human GSK3B in complex with ARN1484 | | Descriptor: | (3S)-1-[(2-fluorophenoxy)acetyl]-N-(pyridin-2-yl)pyrrolidine-3-carboxamide, CHLORIDE ION, Glycogen synthase kinase-3 beta | | Authors: | Tripathi, S.K, Balboni, B, Giabbai, B, Storici, P, Girotto, S, Cavalli, A. | | Deposit date: | 2022-02-25 | | Release date: | 2022-04-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Identification of Novel GSK-3 beta Hits Using Competitive Biophysical Assays.
Int J Mol Sci, 23, 2022
|
|
7YAQ
 
 | | Phloem lectin (PP2)C34S mutant | | Descriptor: | 17 kDa phloem lectin | | Authors: | Sivaji, N, Bobbili, K.B, Priya, B, Suguna, K, Surolia, A. | | Deposit date: | 2022-06-28 | | Release date: | 2023-03-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.853 Å) | | Cite: | Structure and interactions of the phloem lectin (phloem protein 2) Cus17 from Cucumis sativus.
Structure, 31, 2023
|
|
6OOT
 
 | | HIV-1 Protease NL4-3 L89V, L90M Mutant in complex with darunavir | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, NL4-3 PROTEASE, SULFATE ION | | Authors: | Henes, M, Kosovrasti, K, Lockbaum, G.J, Leidner, F, Nachum, G.S, Nalivaika, E.A, Bolon, D.N.A, KurtYilmaz, N, Schiffer, C.A, Whitfield, T.W. | | Deposit date: | 2019-04-23 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.822 Å) | | Cite: | Molecular Determinants of Epistasis in HIV-1 Protease: Elucidating the Interdependence of L89V and L90M Mutations in Resistance.
Biochemistry, 58, 2019
|
|
5MCI
 
 | | Radiation damage to GH7 Family Cellobiohydrolase from Daphnia pulex: Dose (DWD) 11.9 MGy | | Descriptor: | Cellobiohydrolase CHBI, GLYCEROL, SULFATE ION | | Authors: | Bury, C.S, McGeehan, J.E, Ebrahim, A, Garman, E.F. | | Deposit date: | 2016-11-09 | | Release date: | 2017-01-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | OH cleavage from tyrosine: debunking a myth.
J Synchrotron Radiat, 24, 2017
|
|
