8EMZ
 
 | | Structure of GII.17 norovirus in complex with Nanobody 2 | | Descriptor: | 1,2-ETHANEDIOL, GII.17 P domain, Nanobody 2 | | Authors: | Kher, G, Sabin, C, Pancera, M, Koromyslova, A, Hansman, G. | | Deposit date: | 2022-09-28 | | Release date: | 2023-03-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Direct Blockade of the Norovirus Histo-Blood Group Antigen Binding Pocket by Nanobodies.
J.Virol., 97, 2023
|
|
5MY7
 
 | | Adhesin Complex Protein from Neisseria meningitidis | | Descriptor: | Adhesin, DI(HYDROXYETHYL)ETHER, SODIUM ION | | Authors: | Derrick, J.P, Awanye, A. | | Deposit date: | 2017-01-25 | | Release date: | 2017-06-21 | | Last modified: | 2017-07-12 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of the Neisseria Adhesin Complex Protein (ACP) and its role as a novel lysozyme inhibitor.
PLoS Pathog., 13, 2017
|
|
5JQT
 
 | | Crystal structure of human carbonic anhydrase II in complex with Benzoxaborole at pH 7.4 | | Descriptor: | 1,1-dihydroxy-1,3-dihydro-2,1-benzoxaborol-1-ium, 2,1-benzoxaborol-1(3H)-ol, 4-(HYDROXYMERCURY)BENZOIC ACID, ... | | Authors: | Alterio, V, Esposito, D, Di Fiore, A, De Simone, G. | | Deposit date: | 2016-05-05 | | Release date: | 2016-10-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Benzoxaborole as a new chemotype for carbonic anhydrase inhibition.
Chem.Commun.(Camb.), 52, 2016
|
|
8HFH
 
 | | CENP-E motor domain in complex with AMPPNP and Mg2+ | | Descriptor: | Centromere-associated protein E, IMIDAZOLE, MAGNESIUM ION, ... | | Authors: | Shibuya, A, Yokoyama, H. | | Deposit date: | 2022-11-10 | | Release date: | 2023-03-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the motor domain of centromere-associated protein E in complex with a non-hydrolysable ATP analogue.
Febs Lett., 597, 2023
|
|
6EYN
 
 | | Structure of the 8D6 (anti-IgE) Fab | | Descriptor: | 1,2-ETHANEDIOL, 8D6 Fab heavy chain, 8D6 Fab light chain, ... | | Authors: | Chen, J.B, Ramadani, F, Pang, M.O.Y, Beavil, R.L, Holdom, M.D, Mitropoulou, A.N, Beavil, A.J, Gould, H.J, Chang, T.W, Sutton, B.J, McDonnell, J.M, Davies, A.M. | | Deposit date: | 2017-11-13 | | Release date: | 2018-08-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for selective inhibition of immunoglobulin E-receptor interactions by an anti-IgE antibody.
Sci Rep, 8, 2018
|
|
7YX5
 
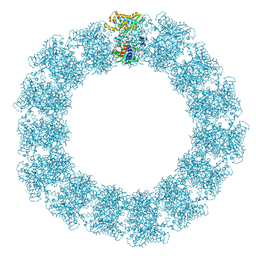 | | Structure of the Mimivirus genomic fibre in its relaxed 5-start helix form | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Putative glucose-methanol-choline oxidoreductase protein | | Authors: | Villalta, A, Schmitt, A, Estrozi, L.F, Quemin, E.R.J, Alempic, J.M, Lartigue, A, Prazak, V, Belmudes, L, Vasishtan, D, Colmant, A.M.G, Honore, F.A, Coute, Y, Grunewald, K, Abergel, C. | | Deposit date: | 2022-02-15 | | Release date: | 2022-08-10 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | The giant mimivirus 1.2 Mb genome is elegantly organized into a 30 nm diameter helical protein shield.
Elife, 11, 2022
|
|
5M6D
 
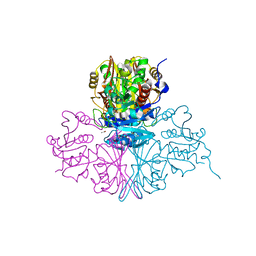 | | Streptococcus pneumoniae Glyceraldehyde-3-Phosphate Dehydrogenase (SpGAPDH) crystal structure | | Descriptor: | ACETIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Gaboriaud, C, Moreau, C.P, Di Guilmi, A.M. | | Deposit date: | 2016-10-25 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Deciphering Key Residues Involved in the Virulence-promoting Interactions between Streptococcus pneumoniae and Human Plasminogen.
J. Biol. Chem., 292, 2017
|
|
7S2I
 
 | | Crystal structure of sulfonamide resistance enzyme Sul1 in complex with 6-hydroxymethylpterin | | Descriptor: | 6-HYDROXYMETHYLPTERIN, CHLORIDE ION, GLYCEROL, ... | | Authors: | Stogios, P.J, Skarina, T, Kim, Y, Venkatesan, M, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID), Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-09-03 | | Release date: | 2023-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Molecular mechanism of plasmid-borne resistance to sulfonamide antibiotics.
Nat Commun, 14, 2023
|
|
5M6S
 
 | |
6S3Q
 
 | | Structure of human excitatory amino acid transporter 3 (EAAT3) in complex with TFB-TBOA | | Descriptor: | (2~{S},3~{S})-2-azanyl-3-[[3-[[4-(trifluoromethyl)phenyl]carbonylamino]phenyl]methoxy]butanedioic acid, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Baronina, A, Pike, A.C.W, Yu, X, Dong, Y.Y, Shintre, C.A, Tessitore, A, Chu, A, Rotty, B, Venkaya, S, Mukhopadhyay, S, Borkowska, O, Chalk, R, Shrestha, L, Burgess-Brown, N.A, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Han, S, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-06-25 | | Release date: | 2020-07-08 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Structure of human excitatory amino acid transporter 3 (EAAT3)
TO BE PUBLISHED
|
|
7S2L
 
 | | Crystal structure of sulfonamide resistance enzyme Sul3 apoenzyme | | Descriptor: | CHLORIDE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Stogios, P.J, Venkatesan, M, Michalska, K, Mesa, N, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID), Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-09-03 | | Release date: | 2023-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Molecular mechanism of plasmid-borne resistance to sulfonamide antibiotics.
Nat Commun, 14, 2023
|
|
5JPC
 
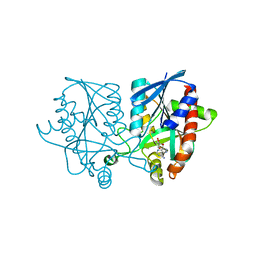 | | Joint X-ray/neutron structure of MTAN complex with Formycin A | | Descriptor: | (1S)-1-(7-amino-1H-pyrazolo[4,3-d]pyrimidin-3-yl)-1,4-anhydro-D-ribitol, Aminodeoxyfutalosine nucleosidase | | Authors: | Banco, M.T, Kovalevsky, A.Y, Ronning, D.R. | | Deposit date: | 2016-05-03 | | Release date: | 2016-11-16 | | Last modified: | 2024-03-06 | | Method: | NEUTRON DIFFRACTION (2.5 Å), X-RAY DIFFRACTION | | Cite: | Neutron structures of the Helicobacter pylori 5'-methylthioadenosine nucleosidase highlight proton sharing and protonation states.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
7YX4
 
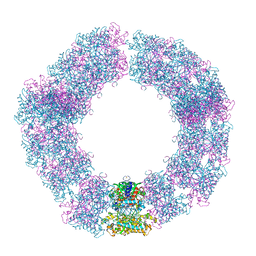 | | Structure of the Mimivirus genomic fibre in its compact 5-start helix form | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Putative glucose-methanol-choline oxidoreductase protein | | Authors: | Villalta, A, Schmitt, A, Estrozi, L.F, Quemin, E.R.J, Alempic, J.M, Lartigue, A, Prazak, V, Belmudes, L, Vasishtan, D, Colmant, A.M.G, Honore, F.A, Coute, Y, Grunewald, K, Abergel, C. | | Deposit date: | 2022-02-15 | | Release date: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | The giant mimivirus 1.2 Mb genome is elegantly organized into a 30 nm diameter helical protein shield.
Elife, 11, 2022
|
|
6S4L
 
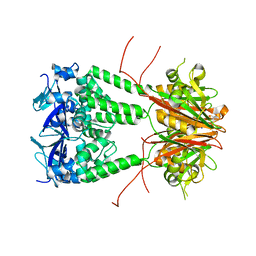 | | Structure of human KCTD1 | | Descriptor: | BTB/POZ domain-containing protein KCTD1, IODIDE ION, SODIUM ION | | Authors: | Pinkas, D.M, Bufton, J.C, Fox, A.E, Pike, A.C.W, Newman, J.A, Krojer, T, Shrestha, L, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C, Edwards, A, Bountra, C, Bullock, A.N. | | Deposit date: | 2019-06-28 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structure of human KCTD1
To be published
|
|
6MWD
 
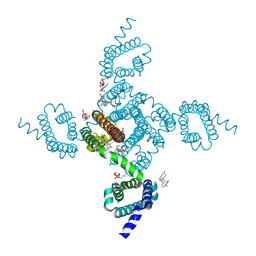 | | NavAb Voltage-gated Sodium Channel, residues 1-239 with mutation T206S | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, ACETATE ION, ... | | Authors: | Lenaeus, M.J, Catterall, W.A. | | Deposit date: | 2018-10-29 | | Release date: | 2018-12-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.327 Å) | | Cite: | Molecular dissection of multiphase inactivation of the bacterial sodium channel NaVAb.
J. Gen. Physiol., 151, 2019
|
|
5JSH
 
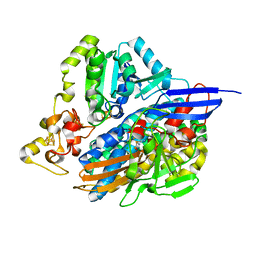 | | The 3D structure of recombinant [NiFeSe] hydrogenase from Desulfovibrio Vulgaris Hildenborough in the oxidized state at 1.30 Angstrom | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, CHLORIDE ION, FE (II) ION, ... | | Authors: | Marques, M.C, Pereira, I.A.C, Matias, P.M. | | Deposit date: | 2016-05-08 | | Release date: | 2017-03-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The direct role of selenocysteine in [NiFeSe] hydrogenase maturation and catalysis.
Nat. Chem. Biol., 13, 2017
|
|
6MX5
 
 | |
7PL3
 
 | |
7YX3
 
 | | Structure of the Mimivirus genomic fibre in its compact 6-start helix form | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Putative GMC-type oxidoreductase | | Authors: | Villalta, A, Schmitt, A, Estrozi, L.F, Quemin, E.R.J, Alempic, J.M, Lartigue, A, Prazak, V, Belmudes, L, Vasishtan, D, Colmant, A.M.G, Honore, F.A, Coute, Y, Grunewald, K, Abergel, C. | | Deposit date: | 2022-02-15 | | Release date: | 2022-08-10 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | The giant mimivirus 1.2 Mb genome is elegantly organized into a 30-nm diameter helical protein shield.
Elife, 11, 2022
|
|
6X63
 
 | | Atomic-Resolution Structure of HIV-1 Capsid Tubes by Magic Angle Spinning NMR | | Descriptor: | HIV-1 capsid protein | | Authors: | Lu, M, Russell, R.W, Bryer, A, Quinn, C.M, Hou, G, Zhang, H, Schwieters, C.D, Perilla, J.R, Gronenborn, A.M, Polenova, T. | | Deposit date: | 2020-05-27 | | Release date: | 2020-09-02 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Atomic-resolution structure of HIV-1 capsid tubes by magic-angle spinning NMR.
Nat.Struct.Mol.Biol., 27, 2020
|
|
5MKA
 
 | | Maltodextrin binding protein MalE1 from L. casei BL23 bound to gamma-cyclodextrin | | Descriptor: | Cyclooctakis-(1-4)-(alpha-D-glucopyranose), MalE1 | | Authors: | Homburg, C, Bommer, M, Wuttge, S, Hobe, C, Beck, S, Dobbek, H, Deutscher, J, Licht, A, Schneider, E. | | Deposit date: | 2016-12-02 | | Release date: | 2017-07-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.149 Å) | | Cite: | Inducer exclusion in Firmicutes: insights into the regulation of a carbohydrate ATP binding cassette transporter from Lactobacillus casei BL23 by the signal transducing protein P-Ser46-HPr.
Mol. Microbiol., 105, 2017
|
|
5JVE
 
 | |
7TAA
 
 | | FAMILY 13 ALPHA AMYLASE IN COMPLEX WITH ACARBOSE | | Descriptor: | CALCIUM ION, MODIFIED ACARBOSE HEXASACCHARIDE, TAKA AMYLASE | | Authors: | Davies, G.J, Brzozowski, A.M. | | Deposit date: | 1997-10-06 | | Release date: | 1998-11-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure of the Aspergillus oryzae alpha-amylase complexed with the inhibitor acarbose at 2.0 A resolution.
Biochemistry, 36, 1997
|
|
5JVJ
 
 | | C4-type pyruvate phosphate dikinase: different conformational states of the nucleotide binding domain in the dimer | | Descriptor: | MAGNESIUM ION, PHOSPHOENOLPYRUVATE, Pyruvate, ... | | Authors: | Minges, A, Hoeppner, A, Groth, G. | | Deposit date: | 2016-05-11 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.898 Å) | | Cite: | Structural intermediates and directionality of the swiveling motion of Pyruvate Phosphate Dikinase.
Sci Rep, 7, 2017
|
|
6S7T
 
 | | Cryo-EM structure of human oligosaccharyltransferase complex OST-B | | Descriptor: | (2Z,6Z,10Z,14Z,18Z,22Z,26Z)-3,7,11,15,19,23,27,31-octamethyldotriaconta-2,6,10,14,18,22,26,30-octaen-1-yl dihydrogen phosphate, (2~{S},3~{R},4~{R},5~{S},6~{S})-2-(hydroxymethyl)-6-[(1~{S},2~{R},3~{R},4~{R},5'~{S},6~{S},7~{R},8~{S},9~{R},12~{R},13~{R},15~{S},16~{S},18~{R})-5',7,9,13-tetramethyl-3,15-bis(oxidanyl)spiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icosane-6,2'-oxane]-16-yl]oxy-oxane-3,4,5-triol, (4R,7R)-4-hydroxy-N,N,N-trimethyl-4,9-dioxo-7-[(undecanoyloxy)methyl]-3,5,8-trioxa-4lambda~5~-phosphadocosan-1-aminium, ... | | Authors: | Ramirez, A.S, Kowal, J, Locher, K.P. | | Deposit date: | 2019-07-05 | | Release date: | 2019-12-18 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-electron microscopy structures of human oligosaccharyltransferase complexes OST-A and OST-B.
Science, 366, 2019
|
|
