7R3B
 
 | | S-adenosylmethionine synthetase from Lactobacillus plantarum complexed with AMPPNP, methionine and SAM | | Descriptor: | (DIPHOSPHONO)AMINOPHOSPHONIC ACID, MAGNESIUM ION, PHENYLALANINE, ... | | Authors: | Shahar, A, Kleiner, D, Bershtein, S, Zarivach, R. | | Deposit date: | 2022-02-07 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Evolution of homo-oligomerization of methionine S-adenosyltransferases is replete with structure-function constrains.
Protein Sci., 31, 2022
|
|
6L5Y
 
 | | Carbonmonoxy human hemoglobin A in the R2 quaternary structure at 140 K: Light (2 min) | | Descriptor: | CARBON MONOXIDE, Hemoglobin subunit alpha, Hemoglobin subunit beta, ... | | Authors: | Shibayama, N, Park, S.Y, Ohki, M, Sato-Tomita, A. | | Deposit date: | 2019-10-24 | | Release date: | 2020-02-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Direct observation of ligand migration within human hemoglobin at work.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8SUZ
 
 | | Open State of the SARS-CoV-2 Envelope Protein Transmembrane Domain, Determined by Solid-State NMR | | Descriptor: | Envelope small membrane protein | | Authors: | Medeiros-Silva, J, Dregni, A.J, Somberg, N.H, Hong, M. | | Deposit date: | 2023-05-14 | | Release date: | 2023-10-25 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Atomic structure of the open SARS-CoV-2 E viroporin.
Sci Adv, 9, 2023
|
|
4W9E
 
 | | pVHL:EloB:EloC in complex with (2S,4R)-1-(3,3-dimethylbutanoyl)-4-hydroxy-N-(4-(thiazol-5-yl)benzyl)pyrrolidine-2-carboxamide (ligand 4) | | Descriptor: | (4R)-1-(3,3-dimethylbutanoyl)-4-hydroxy-N-[4-(1,3-thiazol-5-yl)benzyl]-L-prolinamide, Transcription elongation factor B polypeptide 1, Transcription elongation factor B polypeptide 2, ... | | Authors: | Gadd, M.S, Soares, P, Galdeano, C, van Molle, I, Ciulli, A. | | Deposit date: | 2014-08-27 | | Release date: | 2014-09-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-Guided Design and Optimization of Small Molecules Targeting the Protein-Protein Interaction between the von Hippel-Lindau (VHL) E3 Ubiquitin Ligase and the Hypoxia Inducible Factor (HIF) Alpha Subunit with in Vitro Nanomolar Affinities.
J.Med.Chem., 57, 2014
|
|
2GHO
 
 | | Recombinant Thermus aquaticus RNA polymerase for Structural Studies | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta',DNA-directed RNA polymerase subunit beta' | | Authors: | Lamour, V, Darst, S.A. | | Deposit date: | 2006-03-27 | | Release date: | 2006-05-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (5 Å) | | Cite: | Recombinant Thermus aquaticus RNA Polymerase for Structural Studies.
J.Mol.Biol., 359, 2006
|
|
2FSS
 
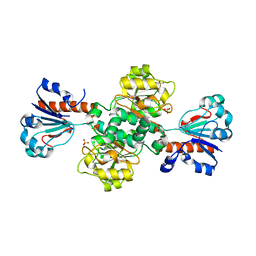 | |
4FCU
 
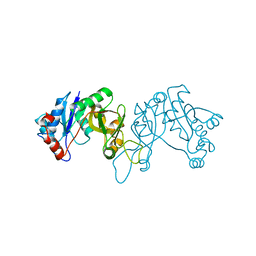 | | 1.9 Angstrom Crystal Structure of 3-deoxy-manno-octulosonate Cytidylyltransferase (kdsB) from Acinetobacter baumannii without His-Tag Bound to the Active Site | | Descriptor: | 3-deoxy-manno-octulosonate cytidylyltransferase | | Authors: | Minasov, G, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-05-25 | | Release date: | 2012-06-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 1.9 Angstrom Crystal Structure of 3-deoxy-manno-octulosonate Cytidylyltransferase (kdsB) from Acinetobacter baumannii without His-Tag Bound to the Active Site.
TO BE PUBLISHED
|
|
6KSS
 
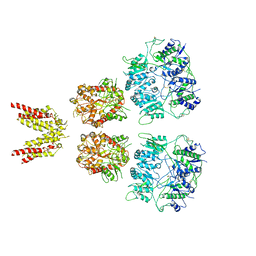 | |
4W9F
 
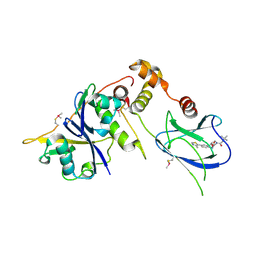 | | pVHL:EloB:EloC in complex with (2S,4R)-1-(3,3-dimethylbutanoyl)-4-hydroxy-N-(4-(4-methylthiazol-5-yl)benzyl)pyrrolidine-2-carboxamide (ligand 5) | | Descriptor: | (4R)-1-(3,3-dimethylbutanoyl)-4-hydroxy-N-[4-(4-methyl-1,3-thiazol-5-yl)benzyl]-L-prolinamide, Transcription elongation factor B polypeptide 1, Transcription elongation factor B polypeptide 2, ... | | Authors: | van Molle, I, Hewitt, S, Galdeano, C, Gadd, M.S, Ciulli, A. | | Deposit date: | 2014-08-27 | | Release date: | 2014-09-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Guided Design and Optimization of Small Molecules Targeting the Protein-Protein Interaction between the von Hippel-Lindau (VHL) E3 Ubiquitin Ligase and the Hypoxia Inducible Factor (HIF) Alpha Subunit with in Vitro Nanomolar Affinities.
J.Med.Chem., 57, 2014
|
|
8RNW
 
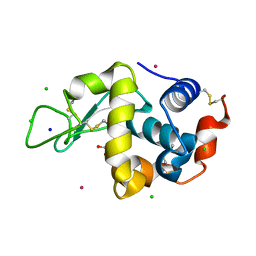 | | Hen Egg White Lysozyme soaked with trans-Ru(DMSO)4Cl2 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C, ... | | Authors: | Oszajca, M, Flejszar, M, Szura, A, Drozdz, P, Brindell, M, Kurpiewska, K. | | Deposit date: | 2024-01-11 | | Release date: | 2024-01-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Exploring the coordination chemistry of ruthenium complexes with lysozymes: structural and in-solution studies.
Front Chem, 12, 2024
|
|
4W9X
 
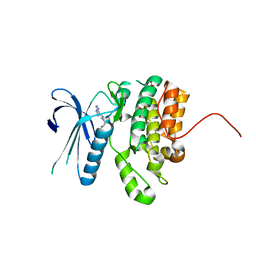 | | Crystal Structure of BMP-2-inducible kinase in complex with baricitinib | | Descriptor: | 1,2-ETHANEDIOL, BMP-2-inducible protein kinase, Baricitinib | | Authors: | Sorrell, F.J, Elkins, J.M, Krojer, T, Williams, E, Savitsky, P, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-08-28 | | Release date: | 2014-09-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Family-wide Structural Analysis of Human Numb-Associated Protein Kinases.
Structure, 24, 2016
|
|
8RNX
 
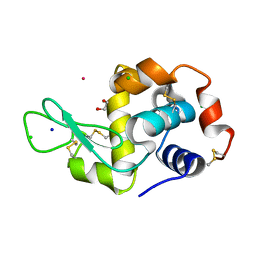 | | Hen Egg White Lysozyme soaked with [HIsq][trans-RuCl4(DMSO)(Isq)] | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C, ... | | Authors: | Oszajca, M, Flejszar, M, Szura, A, Drozdz, P, Brindell, M, Kurpiewska, K. | | Deposit date: | 2024-01-11 | | Release date: | 2024-01-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Exploring the coordination chemistry of ruthenium complexes with lysozymes: structural and in-solution studies.
Front Chem, 12, 2024
|
|
8RNV
 
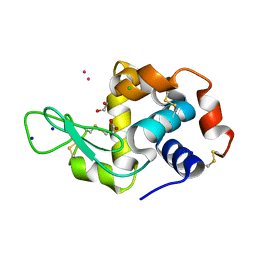 | | Hen Egg White Lysozyme soaked with cis-Ru(DMSO)4Cl2 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Oszajca, M, Flejszar, M, Szura, A, Drozdz, P, Brindell, M, Kurpiewska, K. | | Deposit date: | 2024-01-11 | | Release date: | 2024-01-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Exploring the coordination chemistry of ruthenium complexes with lysozymes: structural and in-solution studies.
Front Chem, 12, 2024
|
|
5KT0
 
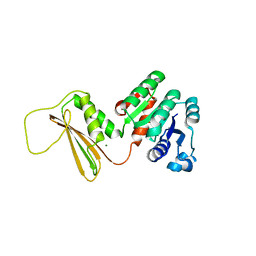 | | Dihydrodipicolinate reductase from the industrial and evolutionarily important cyanobacteria Anabaena variabilis. | | Descriptor: | 4-hydroxy-tetrahydrodipicolinate reductase, MAGNESIUM ION | | Authors: | Christensen, J.B, Soares da Costa, T.P, Faou, P, Pearce, F.G, Panjikar, S, Perugini, M.A. | | Deposit date: | 2016-07-10 | | Release date: | 2016-09-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Dihydrodipicolinate Reductase from the industrial and evolutionarily important cyanobacteria Anabaena variabilis.
To Be Published
|
|
4F9N
 
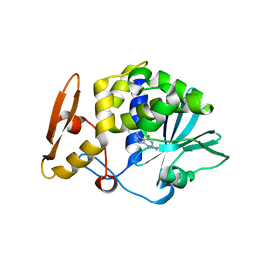 | | Crystal structure of the complex of type I Ribosome inactivating protein from Momordica balsamina with N7-methylated guanine at 2.65 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-amino-7-methyl-1,7-dihydro-6H-purin-6-one, Ribosome inactivating protein | | Authors: | Yamini, S, Kushwaha, G.S, Bhushan, A, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2012-05-19 | | Release date: | 2012-06-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structure of the complex of type I Ribosome inactivating protein from Momordica balsamina with N7-methylated guanine at 2.65 A resolution
To be Published
|
|
7RR7
 
 | | Multidrug efflux pump subunit AcrB | | Descriptor: | DODECANE, Multidrug efflux pump subunit AcrB, PHOSPHATIDYLETHANOLAMINE | | Authors: | Trinh, T.K.H, Cabezas, A, Catalano, C, Qiu, W, des Georges, A, Guo, Y. | | Deposit date: | 2021-08-09 | | Release date: | 2022-08-17 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Multidrug efflux pump subunit AcrB
To Be Published
|
|
7RR6
 
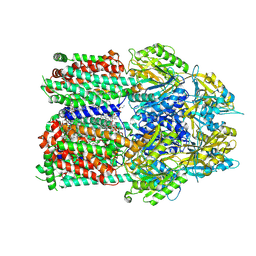 | | Multidrug efflux pump subunit AcrB | | Descriptor: | DODECANE, Multidrug efflux pump subunit AcrB, PHOSPHATIDYLETHANOLAMINE | | Authors: | Trinh, T.K.H, Cabezas, A, Catalano, C, Qiu, W, des Georges, A, Guo, Y. | | Deposit date: | 2021-08-09 | | Release date: | 2022-08-17 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Multidrug efflux pump subunit AcrB
To Be Published
|
|
7RR8
 
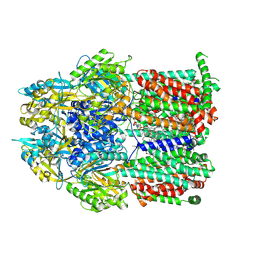 | | Multidrug efflux pump subunit AcrB | | Descriptor: | DODECANE, Multidrug efflux pump subunit AcrB, PHOSPHATIDYLETHANOLAMINE | | Authors: | Trinh, T.K.H, Cabezas, A, Catalano, C, Qiu, W, des Georges, A, Guo, Y. | | Deposit date: | 2021-08-09 | | Release date: | 2022-08-17 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.51 Å) | | Cite: | Multidrug efflux pump subunit AcrB
To Be Published
|
|
6LFH
 
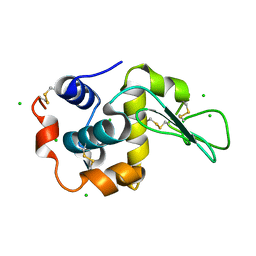 | |
4FBJ
 
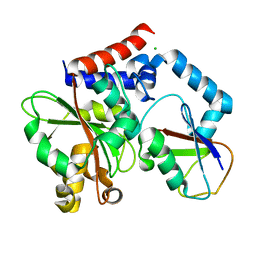 | |
8RNY
 
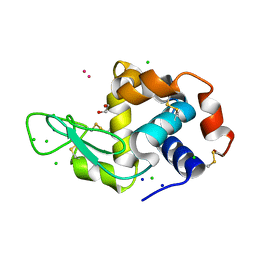 | | Hen Egg White Lysozyme soaked with with [H2Ind][trans-RuCl4(DMSO)(HInd)] | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C, ... | | Authors: | Oszajca, M, Flejszar, M, Szura, A, Drozdz, P, Brindell, M, Kurpiewska, K. | | Deposit date: | 2024-01-11 | | Release date: | 2024-02-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Exploring the coordination chemistry of ruthenium complexes with lysozymes: structural and in-solution studies.
Front Chem, 12, 2024
|
|
4W9J
 
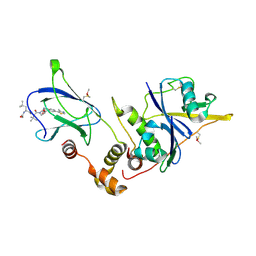 | | pVHL:EloB:EloC in complex with (2S,4R)-1-((S)-2-((S)-2-acetamido-4-methylpentanamido)-3,3-dimethylbutanoyl)-4-hydroxy-N-(4-(4-methylthiazol-5-yl)benzyl)pyrrolidine-2-carboxamide (ligand 13) | | Descriptor: | N-acetyl-L-leucyl-3-methyl-L-valyl-(4R)-4-hydroxy-N-[4-(4-methyl-1,3-thiazol-5-yl)benzyl]-L-prolinamide, Transcription elongation factor B polypeptide 1, Transcription elongation factor B polypeptide 2, ... | | Authors: | Gadd, M.S, Galdeano, C, van Molle, I, Ciulli, A. | | Deposit date: | 2014-08-27 | | Release date: | 2014-09-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Guided Design and Optimization of Small Molecules Targeting the Protein-Protein Interaction between the von Hippel-Lindau (VHL) E3 Ubiquitin Ligase and the Hypoxia Inducible Factor (HIF) Alpha Subunit with in Vitro Nanomolar Affinities.
J.Med.Chem., 57, 2014
|
|
4WBT
 
 | | Crystal structure of histidinol-phosphate aminotransferase from Sinorhizobium meliloti in complex with pyridoxal-5'-phosphate | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Shabalin, I.G, Bacal, P, Kowalska, A.K, Cooper, D.R, Stead, M, Hammonds, J, Ahmed, M, Hillerich, B.S, Bonanno, J, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-09-03 | | Release date: | 2014-09-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of histidinol-phosphate aminotransferase from Sinorhizobium meliloti in complex with pyridoxal-5'-phosphate
to be published
|
|
5L8P
 
 | | Thermolysin in complex with JC114 (PEG400 cryo protectant) | | Descriptor: | CALCIUM ION, DIMETHYL SULFOXIDE, N~2~-[(R)-({[(benzyloxy)carbonyl]amino}methyl)(hydroxy)phosphoryl]-N-[(2S)-2,3,3-trimethylbutyl]-L-leucinamide, ... | | Authors: | Krimmer, S.G, Cramer, J, Heine, A, Klebe, G. | | Deposit date: | 2016-06-08 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Rational Design of Thermodynamic and Kinetic Binding Profiles by Optimizing Surface Water Networks Coating Protein-Bound Ligands.
J. Med. Chem., 59, 2016
|
|
7REY
 
 | | MYCOBACTERIUM ABSCESSUS TRNA METHYLTRANSFERASE IN APO FORM | | Descriptor: | SODIUM ION, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Prucha, G.R, Ismail, M, Suske, A, Das, B, Oz, M, Perez, A, Bolen, R, Jayaraman, S, Stojanoff, V, Halloran, J. | | Deposit date: | 2021-07-13 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystal structure of divalent Mg2+ dependent Mycobacterium abscessus tRNA (m1 G37) Methyltransferase (TrmD)
To Be Published
|
|
