5XBJ
 
 | |
8ADW
 
 | | Lipidic alpha-synuclein fibril - polymorph L1C | | Descriptor: | Alpha-synuclein | | Authors: | Frieg, B, Antonschmidt, L, Dienemann, C, Geraets, J.A, Najbauer, E.E, Matthes, D, de Groot, B.L, Andreas, L.B, Becker, S, Griesinger, C, Schroeder, G.F. | | Deposit date: | 2022-07-11 | | Release date: | 2022-10-12 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | The 3D structure of lipidic fibrils of alpha-synuclein.
Nat Commun, 13, 2022
|
|
6UHP
 
 | | Crystal Structure of C148 mGFP-ncDNA-1 | | Descriptor: | C148 mGFP-ncDNA-1 | | Authors: | Winegar, P.W, Hayes, O.G, McMillan, J.R, Figg, C.A, Focia, P.J, Mirkin, C.A. | | Deposit date: | 2019-09-27 | | Release date: | 2020-03-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | DNA-Directed Protein Packing within Single Crystals.
Chem, 6, 2020
|
|
8AOE
 
 | | Specific covalent inhibitor(15) of ERK2 | | Descriptor: | 1,2-ETHANEDIOL, Mitogen-activated protein kinase 1, SULFATE ION, ... | | Authors: | Cleasby, A. | | Deposit date: | 2022-08-08 | | Release date: | 2022-09-28 | | Last modified: | 2023-05-24 | | Method: | X-RAY DIFFRACTION (1.687 Å) | | Cite: | X-ray Screening of an Electrophilic Fragment Library and Application toward the Development of a Novel ERK 1/2 Covalent Inhibitor.
J.Med.Chem., 65, 2022
|
|
4XQW
 
 | | X-ray structure analysis of xylanase-N44E with MES at pH6.0 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Endo-1,4-beta-xylanase 2, IODIDE ION | | Authors: | Wan, Q, Park, J.M, Riccardi, D.M, Hanson, L.B, Fisher, Z, Smith, J.C, Ostermann, A, Schrader, T, Graham, D.E, Coates, L, Langan, P, Kovalevsky, A.Y. | | Deposit date: | 2015-01-20 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Direct determination of protonation states and visualization of hydrogen bonding in a glycoside hydrolase with neutron crystallography.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
7RE3
 
 | | SARS-CoV-2 replication-transcription complex bound to nsp13 helicase - nsp13(2)-RTC dimer | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, CHAPSO, ... | | Authors: | Chen, J, Malone, B, Campbell, E.A, Darst, S.A. | | Deposit date: | 2021-07-12 | | Release date: | 2021-11-24 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Ensemble cryo-EM reveals conformational states of the nsp13 helicase in the SARS-CoV-2 helicase replication-transcription complex.
Nat.Struct.Mol.Biol., 29, 2022
|
|
6I6M
 
 | | Papaver somniferum O-methyltransferase 1 | | Descriptor: | (13aS)-3,10-dimethoxy-5,8,13,13a-tetrahydro-6H-isoquino[3,2-a]isoquinoline-2,9-diol, O-methyltransferase 1, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Cabry, M.P, Offen, W.A, Winzer, T, Li, Y, Graham, I.A, Davies, G.J, Saleh, P. | | Deposit date: | 2018-11-15 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure of Papaver somniferum O-Methyltransferase 1 Reveals Initiation of Noscapine Biosynthesis with Implications for Plant Natural Product Methylation
Acs Catalysis, 2019
|
|
4Y0G
 
 | | beta2 carbohydrate binding module (CBM) of AMP-activated protein kinase (AMPK) | | Descriptor: | 5'-AMP-activated protein kinase subunit beta-2, GLYCEROL | | Authors: | Mobbs, J, Gorman, M.A, Parker, M.W, Gooley, P.R, Griffin, M. | | Deposit date: | 2015-02-06 | | Release date: | 2015-04-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Determinants of oligosaccharide specificity of the carbohydrate-binding modules of AMP-activated protein kinase.
Biochem.J., 468, 2015
|
|
7WNK
 
 | | Crystal structure of a mutant Staphylococcus equorum manganese superoxide dismutase K38R and A121E | | Descriptor: | AZIDE ION, BROMIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Retnoningrum, D.S, Yoshida, H, Artarini, A.A, Ismaya, W.T. | | Deposit date: | 2022-01-18 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Introducing Intermolecular Interaction to Strengthen the Stability of MnSOD Dimer.
Appl.Biochem.Biotechnol., 195, 2023
|
|
7U6V
 
 | | Cryo-EM structure of Shiga toxin 2 in complex with the native ribosomal P-stalk | | Descriptor: | C-terminal domain (CTD) from the Ribosomal P-stalk, Shiga toxin 2a subunit A (Stx2A), Shiga toxin 2a subunit B (Stx2B) | | Authors: | Kulczyk, A.W. | | Deposit date: | 2022-03-06 | | Release date: | 2023-01-11 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM structure of Shiga toxin 2 in complex with the native ribosomal P-stalk reveals residues involved in the binding interaction.
J.Biol.Chem., 299, 2022
|
|
4QRU
 
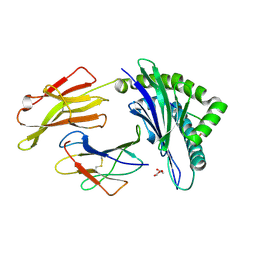 | | Crystal Structure of HLA B*0801 in complex with ELR_MYM, ELRRKMMYM | | Descriptor: | 55 kDa immediate-early protein 1, ACETATE ION, Beta-2-microglobulin, ... | | Authors: | Gras, S, Twist, K.-A, Rossjohn, J. | | Deposit date: | 2014-07-02 | | Release date: | 2015-02-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular imprint of exposure to naturally occurring genetic variants of human cytomegalovirus on the T cell repertoire.
Sci Rep, 4, 2014
|
|
8AEX
 
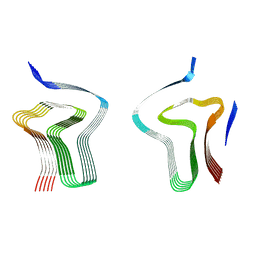 | | Lipidic alpha-synuclein fibril - polymorph L3A | | Descriptor: | Alpha-synuclein | | Authors: | Frieg, B, Antonschmidt, L, Dienemann, C, Geraets, J.A, Najbauer, E.E, Matthes, D, de Groot, B.L, Andreas, L.B, Becker, S, Griesinger, C, Schroeder, G.F. | | Deposit date: | 2022-07-14 | | Release date: | 2022-10-12 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | The 3D structure of lipidic fibrils of alpha-synuclein.
Nat Commun, 13, 2022
|
|
4Y1W
 
 | |
6EC6
 
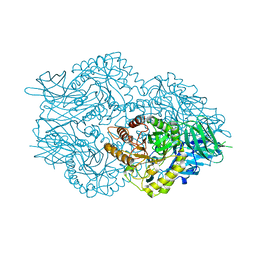 | | Ruminococcus gnavus Beta-glucuronidase | | Descriptor: | Beta-glucuronidase, CHLORIDE ION, GLYCEROL | | Authors: | Biernat, K.A, Redinbo, M.R. | | Deposit date: | 2018-08-07 | | Release date: | 2019-02-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure, function, and inhibition of drug reactivating human gut microbial beta-glucuronidases.
Sci Rep, 9, 2019
|
|
7R9G
 
 | | Catalytically inactive yeast Pseudouridine Synthase, PUS1, bound to RNA | | Descriptor: | CHLORIDE ION, RNA (5'-R(*AP*AP*AP*UP*CP*GP*GP*GP*AP*UP*UP*CP*CP*GP*GP*AP*UP*A)-3'), SULFATE ION, ... | | Authors: | Doyle, L.A, Stoddard, B.L. | | Deposit date: | 2021-06-29 | | Release date: | 2021-12-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structural basis of mRNA recognition and binding by yeast pseudouridine synthase PUS1.
Plos One, 18, 2023
|
|
7A3C
 
 | |
5K28
 
 | | Structure of the unbound SH3 domain of MLK3 | | Descriptor: | Mitogen-activated protein kinase kinase kinase 11 | | Authors: | Kall, S.K, Lavie, A. | | Deposit date: | 2016-05-18 | | Release date: | 2017-12-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Identification of two distinct peptide-binding pockets in the SH3 domain of human mixed-lineage kinase 3.
J. Biol. Chem., 293, 2018
|
|
8ADV
 
 | | Lipidic alpha-synuclein fibril - polymorph L1B | | Descriptor: | Alpha-synuclein | | Authors: | Frieg, B, Antonschmidt, L, Dienemann, C, Geraets, J.A, Najbauer, E.E, Matthes, D, de Groot, B.L, Andreas, L.B, Becker, S, Griesinger, C, Schroeder, G.F. | | Deposit date: | 2022-07-11 | | Release date: | 2022-10-12 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | The 3D structure of lipidic fibrils of alpha-synuclein.
Nat Commun, 13, 2022
|
|
7WNL
 
 | | Crystal structure of a mutant Staphylococcus equorum manganese superoxide dismutase K38R and A121Y | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, AZIDE ION, BROMIDE ION, ... | | Authors: | Retnoningrum, D.S, Yoshida, H, Artarini, A.A, Ismaya, W.T. | | Deposit date: | 2022-01-18 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Introducing Intermolecular Interaction to Strengthen the Stability of MnSOD Dimer.
Appl.Biochem.Biotechnol., 195, 2023
|
|
7R9F
 
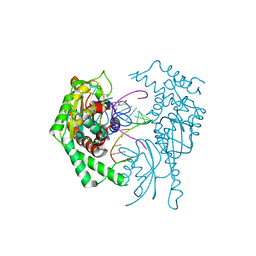 | |
7MJF
 
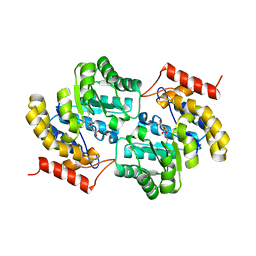 | | Crystal structure of Candidatus Liberibacter solanacearum dihydrodipicolinate synthase with pyruvate and succinic semi-aldehyde bound in active site | | Descriptor: | (4R)-4-oxidanyl-2-oxidanylidene-heptanedioic acid, (4S)-4-hydroxy-2-oxoheptanedioic acid, 4-hydroxy-tetrahydrodipicolinate synthase | | Authors: | Gilkes, J, Frampton, R.A, Board, A.J, Sheen, C.R, Smith, G.R, Dobson, R.C.J. | | Deposit date: | 2021-04-20 | | Release date: | 2021-07-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Candidatus Liberibacter solanacearum dihydrodipicolinate synthase with pyruvate and succinic semi-aldehyde bound in active site
To Be Published
|
|
6RO8
 
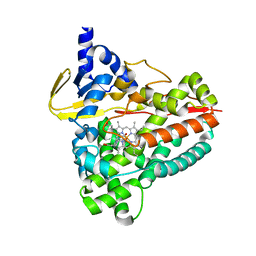 | | The crystal structure of Acinetobacter radioresistens CYP116B5 heme domain | | Descriptor: | Cytochrome P450 RhF, HISTIDINE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Ciaramella, A, Catucci, G, Gilardi, G, Di Nardo, G. | | Deposit date: | 2019-05-10 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of bacterial CYP116B5 heme domain: New insights on class VII P450s structural flexibility and peroxygenase activity.
Int.J.Biol.Macromol., 140, 2019
|
|
7THR
 
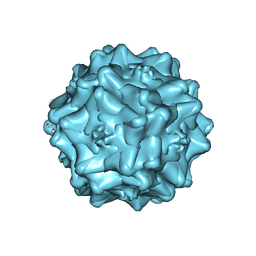 | | Cryo-electron microscopy of Adeno-associated virus serotype 4 at 2.2 A | | Descriptor: | Capsid, MAGNESIUM ION | | Authors: | Zane, G.M, Silveria, M.A, Meyer, N.L, White, T.A, Chapman, M.S. | | Deposit date: | 2022-01-11 | | Release date: | 2023-01-25 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.21 Å) | | Cite: | Cryo-EM structure of adeno-associated virus 4 at 2.2 angstrom resolution.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7RE0
 
 | | SARS-CoV-2 replication-transcription complex bound to nsp13 helicase - nsp13(2)-RTC - swiveled class | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, Helicase, ... | | Authors: | Chen, J, Malone, B, Campbell, E.A, Darst, S.A. | | Deposit date: | 2021-07-12 | | Release date: | 2021-12-01 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Ensemble cryo-EM reveals conformational states of the nsp13 helicase in the SARS-CoV-2 helicase replication-transcription complex.
Nat.Struct.Mol.Biol., 29, 2022
|
|
5JUC
 
 | | Crystal structure of glucosyl-3-phosphoglycerate synthase from Mycobacterium tuberculosis in complex with Mn2+, uridine-diphosphate (UDP) and glucosyl-3-phosphoglycerate (GPG) - GpgS*GPG*UDP*Mn2+_2 | | Descriptor: | (2R)-2-(alpha-D-glucopyranosyloxy)-3-(phosphonooxy)propanoic acid, 1,2-ETHANEDIOL, Glucosyl-3-phosphoglycerate synthase, ... | | Authors: | Albesa-Jove, D, Sancho-Vaello, E, Rodrigo-Unzueta, A, Comino, N, Carreras-Gonzalez, A, Arrasate, P, Urresti, S, Guerin, M.E. | | Deposit date: | 2016-05-10 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Snapshots and Loop Dynamics along the Catalytic Cycle of Glycosyltransferase GpgS.
Structure, 25, 2017
|
|
