7FQH
 
 | | Crystal Structure of human Legumain in complex with (2S)-N-[(3S)-5-amino-5-oxopent-1-yn-3-yl]-1-[1-[4-(cyclopropylmethoxy)phenyl]cyclopropanecarbonyl]pyrrolidine-2-carboxamide | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Legumain, ... | | Authors: | Ehler, A, Benz, J, Bartels, B, Hewings-David, S, Rudolph, M.G. | | Deposit date: | 2022-10-05 | | Release date: | 2023-10-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Crystal Structure of a human Legumain complex
To be published
|
|
5ZBU
 
 | |
5JQF
 
 | | Crystal structure of the lasso peptide Sphingopyxin I (SpI) | | Descriptor: | Sphingopyxin I | | Authors: | Fage, C.D, Hegemann, J.D, Harms, K, Bange, G, Marahiel, M.A. | | Deposit date: | 2016-05-04 | | Release date: | 2016-09-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | Structure and Mechanism of the Sphingopyxin I Lasso Peptide Isopeptidase.
Angew. Chem. Int. Ed. Engl., 55, 2016
|
|
4NR4
 
 | | Crystal structure of the bromodomain of human CREBBP in complex with an isoxazolyl-benzimidazole ligand | | Descriptor: | 1,2-ETHANEDIOL, 1-(4-chlorobenzyl)-5-(3,5-dimethyl-1,2-oxazol-4-yl)-1H-benzimidazole, CREB-binding protein, ... | | Authors: | Filippakopoulos, P, Picaud, S, Felletar, I, Hay, D, Fedorov, O, Martin, S, von Delft, F, Brennan, P, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-11-26 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Crystal structure of the bromodomain of human CREBBP in complex with an isoxazolyl-benzimidazole ligand
TO BE PUBLISHED
|
|
6M44
 
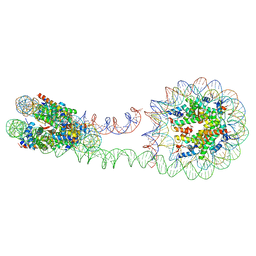 | | 355 bp di-nucleosome harboring cohesive DNA termini (high cryoprotectant) | | Descriptor: | CALCIUM ION, DNA (355-MER), Histone H2A type 1-B/E, ... | | Authors: | Adhireksan, Z, Sharma, D, Lee, P.L, Davey, C.A. | | Deposit date: | 2020-03-05 | | Release date: | 2020-10-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.81 Å) | | Cite: | Near-atomic resolution structures of interdigitated nucleosome fibres.
Nat Commun, 11, 2020
|
|
2Q74
 
 | | Mycobacterium tuberculosis SuhB | | Descriptor: | Inositol-1-monophosphatase | | Authors: | Brown, A.K, Meng, G, Ghadbane, H, Besra, G.S, Futterer, K. | | Deposit date: | 2007-06-06 | | Release date: | 2007-10-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Dimerization of inositol monophosphatase Mycobacterium tuberculosis SuhB is not constitutive, but induced by binding of the activator Mg2+
Bmc Struct.Biol., 7, 2007
|
|
5JRX
 
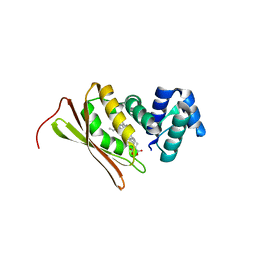 | | Crystal structure of Fe(II) CO-bound H-NOX protein from C. subterraneus | | Descriptor: | CARBON MONOXIDE, Methyl-accepting chemotaxis protein, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Bruegger, J, Hespen, C, Phillips-Piro, C.M, Marletta, M.A. | | Deposit date: | 2016-05-06 | | Release date: | 2016-07-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and Functional Evidence Indicates Selective Oxygen Signaling in Caldanaerobacter subterraneus H-NOX.
Acs Chem.Biol., 11, 2016
|
|
6VPS
 
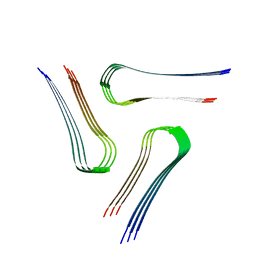 | | Cryo-EM structure of the amyloid core of Drosophila Orb2 isolated from head | | Descriptor: | Translational regulator orb2 | | Authors: | Hervas, R, Rau, M.J, Park, Y, Zhang, W, Murzin, A.G, Fitzpatrick, J.A.J, Scheres, S.H.W, Si, K. | | Deposit date: | 2020-02-04 | | Release date: | 2020-03-18 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Cryo-EM structure of a neuronal functional amyloid implicated in memory persistence in Drosophila
Science, 367, 2020
|
|
4DXE
 
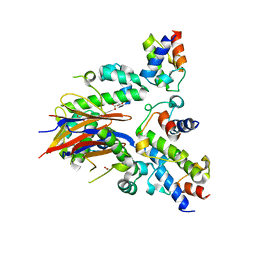 | | 2.52 Angstrom resolution crystal structure of the acyl-carrier-protein synthase (AcpS)-acyl carrier protein (ACP) protein-protein complex from Staphylococcus aureus subsp. aureus COL | | Descriptor: | Acyl carrier protein, MALONATE ION, acyl-carrier-protein synthase | | Authors: | Halavaty, A.S, Minasov, G, Filippova, E.V, Dubrovska, I, Winsor, J, Shuvalova, L, Peterson, S.N, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-02-27 | | Release date: | 2012-03-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | 2.52 Angstrom resolution crystal structure of the acyl-carrier-protein synthase (AcpS)-acyl carrier protein (ACP) protein-protein complex from Staphylococcus aureus subsp. aureus COL
To be Published
|
|
3G7Y
 
 | | Crystal structure of oxidized Ost6L | | Descriptor: | Dolichyl-diphosphooligosaccharide-protein glycosyltransferase subunit OST6 | | Authors: | Stirnimann, C.U, Grimshaw, J.P.A, Schulz, B.L, Brozzo, M.S, Fritsch, F, Glockshuber, R, Capitani, G, Gruetter, M.G, Aebi, M. | | Deposit date: | 2009-02-11 | | Release date: | 2009-06-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.215 Å) | | Cite: | Oxidoreductase activity of oligosaccharyltransferase subunits Ost3p and Ost6p defines site-specific glycosylation efficiency.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3N7Z
 
 | | Crystal structure of acetyltransferase from Bacillus anthracis | | Descriptor: | Acetyltransferase, GNAT family, SODIUM ION | | Authors: | Chang, C, Wu, R, Gornicki, P, Zhang, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-05-27 | | Release date: | 2010-06-16 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Biochemical and Structural Analysis of an Eis Family Aminoglycoside Acetyltransferase from Bacillus anthracis.
Biochemistry, 54, 2015
|
|
1ZGJ
 
 | | Crystal structure of isoflavanone 4'-O-methyltransferase complexed with (+)-pisatin | | Descriptor: | (6AR,12AR)-3-(HYDROXYMETHYL)-6H-[1,3]DIOXOLO[5,6][1]BENZOFURO[3,2-C]CHROMEN-6A(12AH)-OL, Isoflavanone 4'-O-methyltransferase', S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Liu, C.-J, Deavours, B.E, Richard, S, Ferrer, J.-L, Dixon, R.A, Noel, J.P. | | Deposit date: | 2005-04-21 | | Release date: | 2006-08-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for dual functionality of isoflavonoid O-methyltransferases in the evolution of plant defense responses.
Plant Cell, 18, 2006
|
|
1B9L
 
 | | 7,8-DIHYDRONEOPTERIN TRIPHOSPHATE EPIMERASE | | Descriptor: | PROTEIN (EPIMERASE) | | Authors: | Ploom, T, Haussmann, C, Hof, P, Steinbacher, S, Bacher, A, Richardson, J, Huber, R. | | Deposit date: | 1999-02-11 | | Release date: | 2000-02-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of 7,8-dihydroneopterin triphosphate epimerase.
Structure Fold.Des., 7, 1999
|
|
5ZQ5
 
 | | SidE-Ubi | | Descriptor: | SidE, Ubiquitin | | Authors: | Wang, Y, Gao, A, Gao, P. | | Deposit date: | 2018-04-17 | | Release date: | 2018-05-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.487 Å) | | Cite: | Structural Insights into Non-canonical Ubiquitination Catalyzed by SidE.
Cell, 173, 2018
|
|
5KBU
 
 | | Cryo-EM structure of GluA2-2xSTZ complex at 7.8 Angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor 2,Voltage-dependent calcium channel gamma-2 subunit, {[7-morpholin-4-yl-2,3-dioxo-6-(trifluoromethyl)-3,4-dihydroquinoxalin-1(2H)-yl]methyl}phosphonic acid | | Authors: | Twomey, E.C, Yelshanskaya, M.V, Grassucci, R.A, Frank, J, Sobolevsky, A.I. | | Deposit date: | 2016-06-03 | | Release date: | 2016-07-13 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | Elucidation of AMPA receptor-stargazin complexes by cryo-electron microscopy.
Science, 353, 2016
|
|
2G1D
 
 | | Solution Structure of Ribosomal Protein S24E from Thermoplasma acidophilum | | Descriptor: | 30S ribosomal protein S24e | | Authors: | Jeon, B.-Y, Hong, E.-M, Jung, J.-W, Yee, A, Arrowsmith, C.H, Lee, W. | | Deposit date: | 2006-02-14 | | Release date: | 2007-02-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of TA1092, a ribosomal protein S24e from Thermoplasma acidophilum
Proteins, 64, 2006
|
|
6JI1
 
 | |
7KPI
 
 | | Crystal structure of the SPOP MATH domain | | Descriptor: | Speckle-type POZ protein | | Authors: | Usher, E.T, Boal, A.K. | | Deposit date: | 2020-11-11 | | Release date: | 2021-04-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Intrinsically disordered substrates dictate SPOP subnuclear localization and ubiquitination activity.
J.Biol.Chem., 296, 2021
|
|
6TNB
 
 | | X-RAY STRUCTURE OF MPS1 IN COMPLEX WITH COMPOUND 41 | | Descriptor: | (2~{R})-2-(4-fluorophenyl)-~{N}-[4-[2-[(2-methoxy-4-methylsulfonyl-phenyl)amino]-[1,2,4]triazolo[1,5-a]pyridin-6-yl]phenyl]propanamide, CHLORIDE ION, Dual specificity protein kinase TTK | | Authors: | Holton, S.J, Schulze, V.K, Klar, U, Kosemund, D, Siemeister, G, Bader, B, Prechtl, S, Briem, H, Marquardt, T, Schirok, H, Bohlmann, R, Nguyen, D, Fernandez-Montalvan, A, Boemer, U, Eberspaecher, U, Brands, M, Nussbaum, F, Koppitz, M. | | Deposit date: | 2019-12-06 | | Release date: | 2020-05-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Treating Cancer by Spindle Assembly Checkpoint Abrogation: Discovery of Two Clinical Candidates, BAY 1161909 and BAY 1217389, Targeting MPS1 Kinase.
J.Med.Chem., 63, 2020
|
|
5K16
 
 | |
4JVU
 
 | | IgM C2-domain from mouse | | Descriptor: | Ig mu chain C region membrane-bound form | | Authors: | Mueller, R, Graewert, A.M, Kern, T, Madl, T, Peschek, J, Sattler, M, Groll, M, Buchner, J. | | Deposit date: | 2013-03-26 | | Release date: | 2013-06-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | High-resolution structures of the IgM Fc domains reveal principles of its hexamer formation.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6T4Y
 
 | | ROR(gamma)t ligand binding domain in complex with 25-hydroxycholesterol and allosteric ligand MRL871 | | Descriptor: | 25-HYDROXYCHOLESTEROL, 4-{1-[2-chloro-6-(trifluoromethyl)benzoyl]-1H-indazol-3-yl}benzoic acid, GLYCEROL, ... | | Authors: | de Vries, R.M.J.M, Meijer, F.A, Brunsveld, L. | | Deposit date: | 2019-10-15 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Cooperativity between the orthosteric and allosteric ligand binding sites of ROR gamma t.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7W8B
 
 | |
1BAS
 
 | |
6F2S
 
 | | CryoEM structure of Ageratum Yellow Vein virus (AYVV) | | Descriptor: | Capsid protein, coat protein subunit H, coat protein subunit I, ... | | Authors: | Hesketh, E.L, Saunders, K, Fisher, C, Potze, J, Stanley, J, Lomonossoff, G.P, Ranson, N.A. | | Deposit date: | 2017-11-27 | | Release date: | 2018-06-27 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The 3.3 angstrom structure of a plant geminivirus using cryo-EM.
Nat Commun, 9, 2018
|
|
