8OVB
 
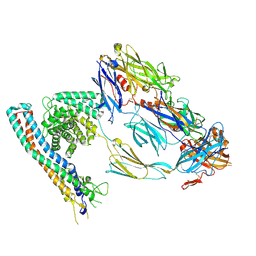 | |
8ZPB
 
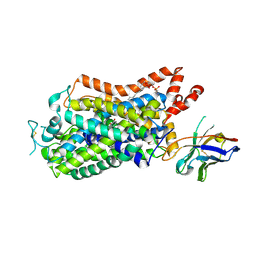 | |
1ZAV
 
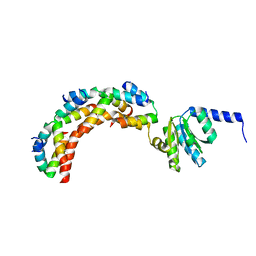 | | Ribosomal Protein L10-L12(NTD) Complex, Space Group P21 | | Descriptor: | 50S ribosomal protein L10, 50S ribosomal protein L7/L12 | | Authors: | Diaconu, M, Kothe, U, Schluenzen, F, Fischer, N, Harms, J.M, Tonevitski, A.G, Stark, H, Rodnina, M.V, Wahl, M.C. | | Deposit date: | 2005-04-07 | | Release date: | 2005-07-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for the Function of the Ribosomal L7/12 Stalk in Factor Binding and GTPase Activation.
Cell(Cambridge,Mass.), 121, 2005
|
|
8OKA
 
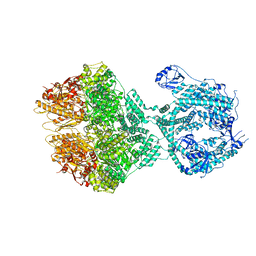 | | Human Mitochondrial Lon Y394F Mutant ADP Bound | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lon protease homolog, mitochondrial | | Authors: | Kereiche, S, Bauer, J.A, Matyas, P, Novacek, J, Kutejova, E. | | Deposit date: | 2023-03-28 | | Release date: | 2024-04-10 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.89 Å) | | Cite: | Polyphosphate and tyrosine phosphorylation in the N-terminal domain of the human mitochondrial Lon protease disrupts its functions.
Sci Rep, 14, 2024
|
|
2XMZ
 
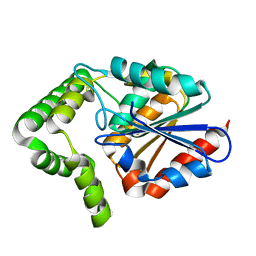 | | Structure of MenH from S. aureus | | Descriptor: | HYDROLASE, ALPHA/BETA HYDROLASE FOLD FAMILY | | Authors: | Dawson, A, Fyfe, P.K, Gillet, F, Hunter, W.N. | | Deposit date: | 2010-07-30 | | Release date: | 2011-05-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Exploiting the High-Resolution Crystal Structure of Staphylococcus Aureus Menh to Gain Insight Into Enzyme Activity.
Bmc Struct.Biol., 11, 2011
|
|
8ONM
 
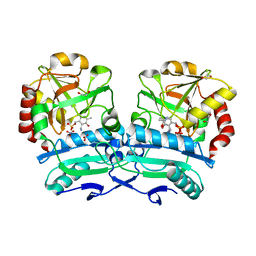 | | Crystal structure of D-amino acid aminotransferase from Aminobacterium colombiense point mutant E113A complexed with D-glutamate | | Descriptor: | (~{Z})-2-[[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylamino]pent-2-enedioic acid, 1,2-ETHANEDIOL, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Matyuta, I.O, Boyko, K.M, Minyaev, M.E, Shilova, S.A, Bezsudnova, E.Y, Popov, V.O. | | Deposit date: | 2023-04-03 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Probing of the structural and catalytic roles of the residues in the active site of transaminase from Aminobacterium colombiense
To Be Published
|
|
6LX9
 
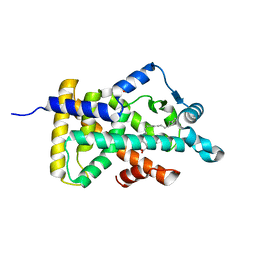 | | X-ray structure of human PPARalpha ligand binding domain-arachidonic acid co-crystals obtained by delipidation and cross-seeding | | Descriptor: | ARACHIDONIC ACID, GLYCEROL, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Saito, K, Honda, A, Ishikawa, R, Oyama, T, Ishii, I. | | Deposit date: | 2020-02-10 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6J9U
 
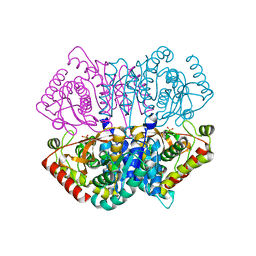 | | Complex structure of Lactobacillus casei lactate dehydrogenase penta mutant with pyruvate | | Descriptor: | L-lactate dehydrogenase, PYRUVIC ACID, SULFATE ION | | Authors: | Arai, K, Miyanaga, A, Uchikoba, H, Fushinobu, S, Taguchi, H. | | Deposit date: | 2019-01-24 | | Release date: | 2019-02-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Crystal structure of penta mutant of L-lactate dehydrogenase from Lactobacillus casei
To Be Published
|
|
7YLO
 
 | | Conversion of indole-3-acetic acid into indole-3-aldehyde in bacteria Metabolic network of tryptophan around the indole-3-aldehyde formation | | Descriptor: | Dyp-type peroxidase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Cheng, J, Luo, Y, Zhu, J, Tan, R, Wang, N, Lebedev, A.A. | | Deposit date: | 2022-07-26 | | Release date: | 2023-07-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Conversion of indole-3-acetic acid into indole-3-aldehyde in bacteria
Metabolic network of tryptophan around the indole-3-aldehyde formation
To Be Published
|
|
6J9X
 
 | | Crystal structure of Trypanosoma brucei gambiense glycerol kinase phosphorylated at Thr12(pyrophosphatase reaction) | | Descriptor: | GLYCEROL, Glycerol kinase | | Authors: | Balogun, E.O, Chishima, T, Ichinose, M, Inaoka, D.K, Kido, Y, Ibrahim, B, Bringaud, F, de Koning, H, McKerrow, J.H, Watanabe, Y, Nozaki, T, Michels, P.A.M, Harada, S, Kita, K, Shiba, T. | | Deposit date: | 2019-01-24 | | Release date: | 2020-01-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Glycerol Kinase of African Human Trypanosomes Possesses a Pyrophosphatase Activity.
To Be Published
|
|
2FB6
 
 | | Structure of Conserved Protein of Unknown Function BT1422 from Bacteroides thetaiotaomicron | | Descriptor: | conserved hypothetical protein | | Authors: | Osipiuk, J, Mulligan, R, Abdullah, J, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-12-08 | | Release date: | 2006-01-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | X-ray crystal structure of conserved hypothetical protein BT1422 from Bacteroides thetaiotaomicron
To be Published
|
|
5JXL
 
 | |
6NIO
 
 | | Crystal Structure of the Molybdate Transporter Periplasmic Protein ModA from Yersinia pestis | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, ACETIC ACID, FORMIC ACID, ... | | Authors: | Kim, Y, Joachimiak, G, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-12-31 | | Release date: | 2019-01-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Crystal Structure of the Molybdate Transporter Periplasmic Protein ModA from Yersinia pestis
To Be Published
|
|
2OLS
 
 | |
2XA8
 
 | | Crystal structure of the Fab domain of omalizumab at 2.41A | | Descriptor: | OMALIZUMAB HEAVY CHAIN, OMALIZUMAB LIGHT CHAIN | | Authors: | Huang, C.H, Hung, F.H.A, Lim, C, Chang, T.W, Ma, C. | | Deposit date: | 2010-03-30 | | Release date: | 2011-05-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structural and Physical Basis for Anti-IgE Therapy.
Sci Rep, 5, 2015
|
|
6JKT
 
 | | Crystal structure of aspartate transcarbamoylase from Trypanosoma cruzi in complex with N-(PHOSPHONACETYL)-L-ASPARTIC ACID (PALA). | | Descriptor: | Aspartate carbamoyltransferase, GLYCEROL, N-(PHOSPHONACETYL)-L-ASPARTIC ACID | | Authors: | Matoba, K, Shiba, T, Nara, T, Aoki, T, Nagasaki, S, Hayamizu, R, Honma, T, Tanaka, A, Inoue, M, Matsuoka, S, Balogun, E.O, Inaoka, D.K, Kita, K, Harada, S. | | Deposit date: | 2019-03-01 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystallographic snapshots of Trypanosoma cruzi aspartate transcarbamoylase
revealed an ordered Bi-Bi reaction mechanism
To Be Published
|
|
6NVF
 
 | | Crystal structure of Mycobacterium tuberculosis dethiobiotin synthetase in complex with fragment analogue 8 | | Descriptor: | (4-{[(1R,2R)-2-(carboxymethyl)cyclopentyl]methyl}phenyl)acetic acid, (4-{[(1S,2S)-2-(carboxymethyl)cyclopentyl]methyl}phenyl)acetic acid, ATP-dependent dethiobiotin synthetase BioD, ... | | Authors: | Thompson, A.P, Polyak, S.W, Wegener, K.L, Bruning, J.B. | | Deposit date: | 2019-02-05 | | Release date: | 2020-02-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.989 Å) | | Cite: | Crystal structure of Mycobacterium tuberculosis dethiobiotin synthetase in complex with fragment analogue 8
To Be Published
|
|
2FPN
 
 | |
2OP6
 
 | | Peptide-binding domain of Heat shock 70 kDa protein D precursor from C.elegans | | Descriptor: | Heat shock 70 kDa protein D | | Authors: | Osipiuk, J, Duggan, E, Gu, M, Voisine, C, Morimoto, R.I, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-01-26 | | Release date: | 2007-02-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | X-ray structure of peptide-binding domain of Heat shock 70 kDa protein D precursor from C.elegans
To be Published
|
|
6W40
 
 | |
2FI9
 
 | | The crystal structure of an outer membrane protein from the Bartonella henselae | | Descriptor: | Outer membrane protein | | Authors: | Zhang, R, Hatzos, C, Clancy, S, Collart, F, Cymborowski, M, Minor, W, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-12-28 | | Release date: | 2006-02-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The 1.8A crystal structure of an outer membrane protein from the Bartonella henselae
To be Published
|
|
2AJL
 
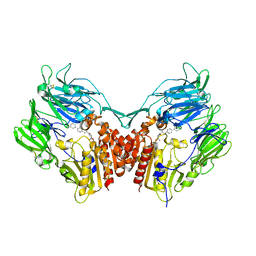 | | X-ray Structure of Novel Biaryl-Based Dipeptidyl peptidase IV inhibitor | | Descriptor: | 1-[2-(S)-AMINO-3-BIPHENYL-4-YL-PROPIONYL]-PYRROLIDINE-2-(S)-CARBONITRILE, 2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl peptidase 4 | | Authors: | Qiao, L, Baumann, C.A, Crysler, C.S, Ninan, N.S, Abad, M.C, Spurlino, J.C, DesJarlais, R.L, Kervinen, J, Neeper, M.P, Bayoumy, S.S. | | Deposit date: | 2005-08-02 | | Release date: | 2005-11-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery, SAR, and X-ray structure of novel biaryl-based dipeptidyl peptidase IV inhibitors
Bioorg.Med.Chem.Lett., 16, 2006
|
|
2OYZ
 
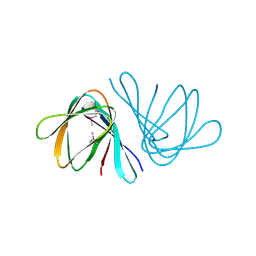 | |
3X3H
 
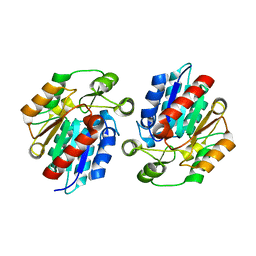 | | Crystal Structure of the Manihot esculenta Hydroxynitrile Lyase (MeHNL) 3KP (K176P, K199P, K224P) triple mutant | | Descriptor: | (S)-hydroxynitrile lyase | | Authors: | Cielo, C.B.C, Yamane, T, Asano, Y, Dadashipour, M, Suzuki, A, Mizushima, T, Komeda, H, Okazaki, S. | | Deposit date: | 2015-01-21 | | Release date: | 2016-03-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Crystallographic Studies of Manihot esculenta hydroxynitrile lyase Lysine-to-Proline mutants
TO BE PUBLISHED
|
|
2FL4
 
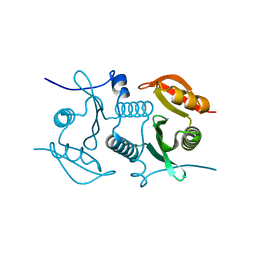 | |
