4IH4
 
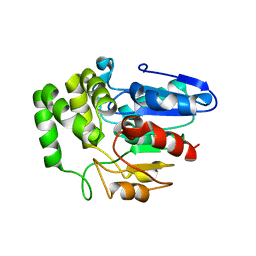 | | Crystal structure of Arabidopsis DWARF14 orthologue, AtD14 | | Descriptor: | AT3g03990/T11I18_10 | | Authors: | Zhou, X.E, Zhao, L.-H, Wu, Z.-S, Yi, W, Li, S, Li, Y, Xu, Y, Xu, T.-H, Liu, Y, Chen, R.-Z, Kovach, A, Kang, Y, Hou, L, He, Y, Zhang, C, Melcher, K, Xu, H.E. | | Deposit date: | 2012-12-18 | | Release date: | 2013-01-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structures of two phytohormone signal-transducing alpha / beta hydrolases: karrikin-signaling KAI2 and strigolactone-signaling DWARF14.
Cell Res., 23, 2013
|
|
8A85
 
 | |
3EUP
 
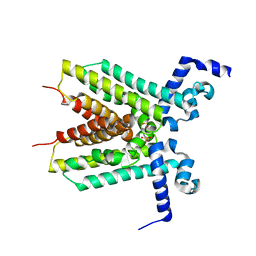 | | The crystal structure of the transcriptional regulator, TetR family from Cytophaga hutchinsonii | | Descriptor: | SULFATE ION, Transcriptional regulator, TetR family | | Authors: | Zhang, R, Bigelow, L, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-10-10 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The crystal structure of the transcriptional regulator, TetR family from Cytophaga hutchinsonii
To be Published
|
|
7ZVE
 
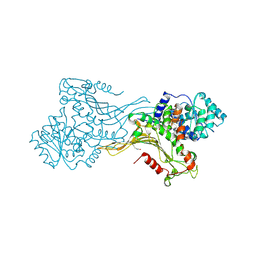 | | K403 acetylated glucose-6-phosphate dehydrogenase (G6PD) | | Descriptor: | COPPER (II) ION, GLYCEROL, Glucose-6-phosphate 1-dehydrogenase | | Authors: | Wu, F, Muskat, N.H, Shahar, A, Arbely, E. | | Deposit date: | 2022-05-15 | | Release date: | 2023-08-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Acetylation-dependent coupling between G6PD activity and apoptotic signaling.
Nat Commun, 14, 2023
|
|
3EZY
 
 | | Crystal structure of probable dehydrogenase TM_0414 from Thermotoga maritima | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, Dehydrogenase | | Authors: | Ramagopal, U.A, Toro, R, Freeman, J, Chang, S, Maletic, M, Gheyi, T, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-10-24 | | Release date: | 2009-01-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Crystal structure of probable dehydrogenase TM_0414 from Thermotoga maritima
To be published
|
|
1TE7
 
 | | Solution NMR Structure of Protein yqfB from Escherichia coli. Northeast Structural Genomics Consortium Target ET99 | | Descriptor: | Hypothetical UPF0267 protein yqfB | | Authors: | Atreya, H.S, Shen, Y, Yee, A, Arrowsmith, C, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-05-24 | | Release date: | 2005-01-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | G-Matrix Fourier Transform NOESY-Based Protocol for High-Quality Protein Structure Determination
J.Am.Chem.Soc., 127, 2005
|
|
3EY5
 
 | | Putative acetyltransferase from GNAT family from Bacteroides thetaiotaomicron. | | Descriptor: | Acetyltransferase-like, GNAT family, SULFATE ION | | Authors: | Osipiuk, J, Bigelow, L, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-10-17 | | Release date: | 2008-11-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | X-ray crystal structure of putative acetyltransferase from GNAT family from Bacteroides thetaiotaomicron.
To be Published
|
|
1T0T
 
 | | Crystallographic structure of a putative chlorite dismutase | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, APC35880, MAGNESIUM ION | | Authors: | Gilski, M, Borek, D, Chen, Y, Collart, F, Joachimiak, A, Otwinowski, Z, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-04-12 | | Release date: | 2004-08-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of APC35880 protein from Bacillus Stearothermophilus
To be Published
|
|
7SJL
 
 | |
6I5J
 
 | | Crystal structure of SOCS2:Elongin C:Elongin B in complex with growth hormone receptor peptide | | Descriptor: | COBALT (II) ION, Elongin-B, Elongin-C, ... | | Authors: | Kung, W.W, Ramachandran, S, Makukhin, N, Bruno, E, Ciulli, A. | | Deposit date: | 2018-11-13 | | Release date: | 2019-05-29 | | Last modified: | 2019-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insights into substrate recognition by the SOCS2 E3 ubiquitin ligase.
Nat Commun, 10, 2019
|
|
2W56
 
 | |
4V2D
 
 | | FLRT2 LRR domain | | Descriptor: | FIBRONECTIN LEUCINE RICH TRANSMEMBRANE PROTEIN 2 | | Authors: | Seiradake, E, del Toro, D, Nagel, D, Cop, F, Haertl, R, Ruff, T, Seyit-Bremer, G, Harlos, K, Border, E.C, Acker-Palmer, A, Jones, E.Y, Klein, R. | | Deposit date: | 2014-10-08 | | Release date: | 2014-11-05 | | Last modified: | 2015-07-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Flrt Structure: Balancing Repulsion and Cell Adhesion in Cortical and Vascular Development
Neuron, 84, 2014
|
|
7SWI
 
 | | cTnC-TnI chimera complexed with A2 | | Descriptor: | 4-(3-cyano-3-methylazetidine-1-carbonyl)-N-[(3R,4S)-7-fluoro-4-hydroxy-6-methyl-3,4-dihydro-2H-1-benzopyran-3-yl]-5-methyl-1H-pyrrole-2-sulfonamide, Troponin C, slow skeletal and cardiac muscles,Troponin I, ... | | Authors: | Poppe, L, Hartman, J.J, Romero, A, Reagan, J.D. | | Deposit date: | 2021-11-19 | | Release date: | 2022-04-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural and Thermodynamic Model for the Activation of Cardiac Troponin.
Biochemistry, 61, 2022
|
|
7SUP
 
 | | NMR structure of cTnC-TnI chimera bound to calcium and A1 | | Descriptor: | 4-(3-cyano-3-methylazetidine-1-carbonyl)-N-[(3S)-7-fluoro-6-methyl-3,4-dihydro-2H-1-benzopyran-3-yl]-5-methyl-1H-pyrrole-2-sulfonamide, CALCIUM ION, Troponin C, ... | | Authors: | Poppe, L, Hartman, J.J, Romero, A, Reagan, J.D. | | Deposit date: | 2021-11-17 | | Release date: | 2022-04-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural and Thermodynamic Model for the Activation of Cardiac Troponin.
Biochemistry, 61, 2022
|
|
7SVC
 
 | | NMR structure of cTnC-TnI chimera bound to calcium and A2 | | Descriptor: | 4-(3-cyano-3-methylazetidine-1-carbonyl)-N-[(3R,4S)-7-fluoro-4-hydroxy-6-methyl-3,4-dihydro-2H-1-benzopyran-3-yl]-5-methyl-1H-pyrrole-2-sulfonamide, CALCIUM ION, Troponin C, ... | | Authors: | Poppe, L, Hartman, J.J, Romero, A, Reagan, J.D. | | Deposit date: | 2021-11-18 | | Release date: | 2022-04-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural and Thermodynamic Model for the Activation of Cardiac Troponin.
Biochemistry, 61, 2022
|
|
6IX6
 
 | | Crystal structure of the complex of peptidyl-tRNA hydrolase with N-propanol at 1.43 A resolution | | Descriptor: | N-PROPANOL, Peptidyl-tRNA hydrolase | | Authors: | Viswanathan, V, Sharma, P, Chaudhary, A, Sharma, S, Singh, T.P. | | Deposit date: | 2018-12-09 | | Release date: | 2018-12-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Crystal structure of the complex of peptidyl-tRNA hydrolase with N-propanol at 1.43 A resolution
To Be Published
|
|
6IXE
 
 | | Crystal structure of SeMet apo SH3BP5 (I41) | | Descriptor: | SH3 domain-binding protein 5, SUCCINIC ACID | | Authors: | Goto-Ito, S, Yamagata, A, Sato, Y, Fukai, S. | | Deposit date: | 2018-12-10 | | Release date: | 2019-03-20 | | Last modified: | 2019-03-27 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Structural basis of guanine nucleotide exchange for Rab11 by SH3BP5.
Life Sci Alliance, 2, 2019
|
|
6J9Q
 
 | | Crystal structure of Trypanosoma brucei gambiense glycerol kinase complex with AMP-PNP. | | Descriptor: | GLYCEROL, Glycerol kinase, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Balogun, E.O, Chishima, T, Ichinose, M, Inaoka, D.K, Kido, Y, Ibrahim, B, de Koning, H, McKerrow, J.H, Watanabe, Y, Nozaki, T, Michels, P.A.M, Harada, S, Kita, K, Shiba, T. | | Deposit date: | 2019-01-24 | | Release date: | 2020-01-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Reaction mechanism of the reverse reaction of African human trypanosomes glycerol kinase.
To Be Published
|
|
6J9S
 
 | | Penta mutant of Lactobacillus casei lactate dehydrogenase | | Descriptor: | GLYCEROL, L-lactate dehydrogenase, SULFATE ION | | Authors: | Arai, K, Miyanaga, A, Uchikoba, H, Fushinobu, S, Taguchi, H. | | Deposit date: | 2019-01-24 | | Release date: | 2019-02-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of penta mutant of L-lactate dehydrogenase from Lactobacillus casei
To Be Published
|
|
7T3J
 
 | | Cryo-EM structure of Csy-AcrIF24 | | Descriptor: | AcrIF24, CRISPR type I-F/YPEST-associated protein Csy2, CRISPR type I-F/YPEST-associated protein Csy3, ... | | Authors: | Mukherjee, I.A, Chang, L. | | Deposit date: | 2021-12-08 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis of AcrIF24 as an anti-CRISPR protein and transcriptional suppressor.
Nat.Chem.Biol., 18, 2022
|
|
2I7R
 
 | |
2I3U
 
 | | Structural studies of protein tyrosine phosphatase beta catalytic domain in complex with inhibitors | | Descriptor: | Receptor-type tyrosine-protein phosphatase beta | | Authors: | Evdokimov, A.G, Pokross, M.E, Walter, R.L, Mekel, M. | | Deposit date: | 2006-08-20 | | Release date: | 2006-08-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Engineering the catalytic domain of human protein tyrosine phosphatase beta for structure-based drug discovery.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2I9D
 
 | |
6WAJ
 
 | | Crystal structure of the UBL domain of human NLE1 | | Descriptor: | NLE1, UNKNOWN ATOM OR ION | | Authors: | Halabelian, L, Zeng, H, Li, Y, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-03-25 | | Release date: | 2020-04-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the UBL domain of human NLE1
To be Published
|
|
2I9T
 
 | | Structure of NF-kB p65-p50 heterodimer bound to PRDII element of B-interferon promoter | | Descriptor: | 5'-D(*AP*GP*TP*GP*GP*GP*AP*AP*AP*TP*TP*CP*CP*TP*CP*TP*G)-3', 5'-D(*CP*AP*GP*AP*GP*GP*AP*AP*TP*TP*TP*CP*CP*CP*AP*CP*T)-3', Nuclear factor NF-kappa-B p105 subunit, ... | | Authors: | Escalante, C.R, Shen, L, Thanos, D, Aggarwal, A.K. | | Deposit date: | 2006-09-06 | | Release date: | 2007-02-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of NF-kappaB p50/p65 heterodimer bound to the PRDII DNA element from the interferon-beta promoter
Structure, 10, 2002
|
|
