7ZTB
 
 | |
6QY2
 
 | | Pink beam serial crystallography: Lysozyme, 5 us exposure, 750 patterns merged | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C, ... | | 著者 | Lieske, J, Tolstikova, A, Meents, A. | | 登録日 | 2019-03-08 | | 公開日 | 2019-09-25 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | 1 kHz fixed-target serial crystallography using a multilayer monochromator and an integrating pixel detector.
Iucrj, 6, 2019
|
|
7PJA
 
 | | Crystal structure of YTHDC1 with compound PSI_DC1_002 | | 分子名称: | 6-methyl-3H-pyridin-2-one, GLYCEROL, SULFATE ION, ... | | 著者 | Bedi, R.K, Huang, D, Caflisch, A. | | 登録日 | 2021-08-23 | | 公開日 | 2021-10-20 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Structure-based design of ligands of the m6A-RNA reader YTHDC1
Eur J Med Chem Rep, 5, 2022
|
|
6QY5
 
 | | Pink beam serial crystallography: Lysozyme, 1 us exposure, 4448 patterns merged (1 chip) | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C, ... | | 著者 | Lieske, J, Tolstikova, A, Meents, A. | | 登録日 | 2019-03-08 | | 公開日 | 2019-10-16 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.84 Å) | | 主引用文献 | 1 kHz fixed-target serial crystallography using a multilayer monochromator and an integrating pixel detector.
Iucrj, 6, 2019
|
|
7PJ7
 
 | |
6PSF
 
 | | Rhinovirus C15 complexed with domains I and II of receptor CDHR3 | | 分子名称: | Cadherin-related family member 3, Capsid protein VP1, Capsid protein VP2, ... | | 著者 | Sun, Y, Watters, K, Klose, T, Palmenberg, A.C. | | 登録日 | 2019-07-12 | | 公開日 | 2020-03-11 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Cryo-EM structure of rhinovirus C15a bound to its cadherin-related protein 3 receptor.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
4X08
 
 | | Structure of H128N/ECP mutant in complex with sulphate anions at 1.34 Angstroms. | | 分子名称: | Eosinophil cationic protein, SULFATE ION | | 著者 | Blanco, J.A, Garcia, J.M, Salazar, V.A, Sanchez, D, Moussauoi, M, Boix, E. | | 登録日 | 2014-11-21 | | 公開日 | 2015-10-07 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.34 Å) | | 主引用文献 | Structure of H128N/ECP mutant in complex with sulphate anions at 1.34 Angstroms.
To Be Published
|
|
5LE5
 
 | | Native human 20S proteasome at 1.8 Angstrom | | 分子名称: | CHLORIDE ION, MAGNESIUM ION, PENTAETHYLENE GLYCOL, ... | | 著者 | Schrader, J, Henneberg, F, Mata, R, Tittmann, K, Schneider, T.R, Stark, H, Bourenkov, G, Chari, A. | | 登録日 | 2016-06-29 | | 公開日 | 2016-08-17 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | The inhibition mechanism of human 20S proteasomes enables next-generation inhibitor design.
Science, 353, 2016
|
|
6FWL
 
 | | Structure of an E333Q variant of the GH99 endo-alpha-mannanase from Bacteroides xylanisolvens in complex alpha-Glc-1,3-(1,2-anhydro-carba-mannose) | | 分子名称: | (1~{S},2~{S},3~{R},4~{R},6~{R})-4-(hydroxymethyl)-7-oxabicyclo[4.1.0]heptane-2,3-diol, ACETATE ION, Glycosyl hydrolase family 71, ... | | 著者 | Sobala, L.F, Speciale, G, Hakki, Z, Fernandes, P.Z, Raich, L, Rojas-Cervellera, V, Bennet, A, Thompson, A.J, Bernardo-Seisdedos, G, Millet, O, Zhu, S, Lu, D, Sollogoub, M, Rovira, C, Jimenez-Barbero, J, Davies, G.J, Williams, S.J. | | 登録日 | 2018-03-06 | | 公開日 | 2019-09-18 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.12 Å) | | 主引用文献 | An Epoxide Intermediate in Glycosidase Catalysis.
Acs Cent.Sci., 6, 2020
|
|
5KCD
 
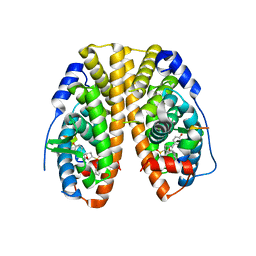 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with an N-methyl Substituted OBHS-N derivative | | 分子名称: | (1S,2R,4S)-5,6-bis(4-hydroxyphenyl)-N-methyl-N-phenyl-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonamide, Estrogen receptor, NCOA2 | | 著者 | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Dharmarajan, V, Goswami, D, Kastrati, I, Novick, S, Nowak, J, Zhou, H.B, Boonmuen, N, Zhao, Y, Min, J, Frasor, J, Katzenellenbogen, B.S, Griffin, P.R, Katzenellenbogen, J.A, Nettles, K.W. | | 登録日 | 2016-06-06 | | 公開日 | 2016-11-16 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.82 Å) | | 主引用文献 | Full antagonism of the estrogen receptor without a prototypical ligand side chain.
Nat. Chem. Biol., 13, 2017
|
|
6NSN
 
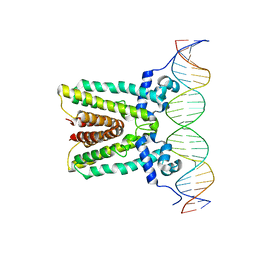 | |
6QMN
 
 | | Crystal structure of a Ribonuclease A-Onconase chimera | | 分子名称: | PHOSPHATE ION, Ribonuclease pancreatic | | 著者 | Esposito, L, Vitagliano, L, Ruggiero, A, Picone, D, Leone, S, Donnarumma, F. | | 登録日 | 2019-02-07 | | 公開日 | 2019-05-08 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.31 Å) | | 主引用文献 | Structure, stability and aggregation propensity of a Ribonuclease A-Onconase chimera.
Int.J.Biol.Macromol., 133, 2019
|
|
5LGB
 
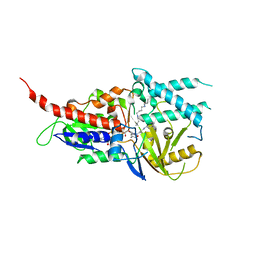 | | Crystal structure of murine N1-acetylpolyamine oxidase in complex with MDL72527 | | 分子名称: | FAD-MDL72527 adduct, N,N'-BIS(2,3-BUTADIENYL)-1,4-BUTANE-DIAMINE, Peroxisomal N(1)-acetyl-spermine/spermidine oxidase,Peroxisomal N(1)-acetyl-spermine/spermidine oxidase | | 著者 | Sjogren, T, Aagaard, A, Wassvik, C, Snijder, A, Barlind, L. | | 登録日 | 2016-07-06 | | 公開日 | 2017-08-16 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | The Structure of Murine N(1)-Acetylspermine Oxidase Reveals Molecular Details of Vertebrate Polyamine Catabolism.
Biochemistry, 56, 2017
|
|
7TIA
 
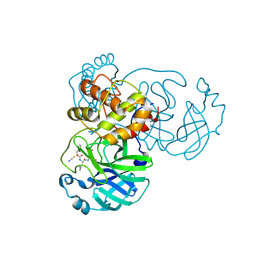 | | Crystal structure of SARS-CoV-2 3CL in complex with inhibitor NK01-14 | | 分子名称: | 3C-like proteinase nsp5, THIOCYANATE ION, benzyl [(2S)-3-cyclopropyl-1-({(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}amino)-1-oxopropan-2-yl]carbamate | | 著者 | Forouhar, F, Liu, H, Iketani, S, Zack, A, Khanizeman, N, Bednarova, E, Fowler, B, Hong, S.J, Mohri, H, Nair, M.S, Huang, Y, Tay, N.E.S, Lee, S, Karan, C, Resnick, S.J, Quinn, C, Li, W, Shion, H, Jurtschenko, C, Lauber, M.A, McDonald, T, Stokes, M.E, Hurst, B, Rovis, T, Chavez, A, Ho, D.D, Stockwell, B.R. | | 登録日 | 2022-01-13 | | 公開日 | 2022-05-04 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.64 Å) | | 主引用文献 | Development of optimized drug-like small molecule inhibitors of the SARS-CoV-2 3CL protease for treatment of COVID-19.
Nat Commun, 13, 2022
|
|
6QZ6
 
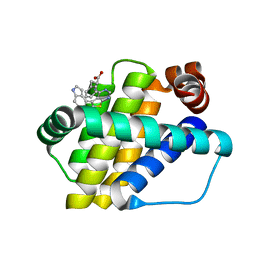 | | Structure of Mcl-1 in complex with compound 8b | | 分子名称: | (2~{R})-2-[[6-ethyl-5-(1~{H}-indol-4-yl)thieno[2,3-d]pyrimidin-4-yl]amino]-3-phenyl-propanoic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | 著者 | Dokurno, P, Szlavik, Z, Ondi, L, Csekei, M, Paczal, A, Szabo, Z.B, Radics, G, Murray, J, Davidson, J, Chen, I, Davis, B, Hubbard, R.E, Pedder, C, Surgenor, A.E, Smith, J, Robertson, A, LeToumelin-Braizat, G, Cauquil, N, Zarka, M, Demarles, D, Perron-Sierra, F, Geneste, O, Kotschy, A. | | 登録日 | 2019-03-11 | | 公開日 | 2019-08-07 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure-Guided Discovery of a Selective Mcl-1 Inhibitor with Cellular Activity.
J.Med.Chem., 62, 2019
|
|
6FXC
 
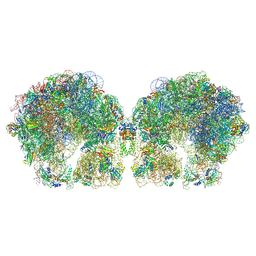 | | The cryo-EM structure of hibernating 100S ribosome dimer from pathogenic Staphylococcus aureus | | 分子名称: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Matzov, D, Aibara, S, Zimmerman, E, Bashan, A, Kidmose, R, Amunts, A, Yonath, A. | | 登録日 | 2018-03-08 | | 公開日 | 2018-03-21 | | 最終更新日 | 2018-11-21 | | 実験手法 | ELECTRON MICROSCOPY (6.76 Å) | | 主引用文献 | The cryo-EM structure of hibernating 100S ribosome dimer from pathogenic Staphylococcus aureus.
Nat Commun, 8, 2017
|
|
6G3K
 
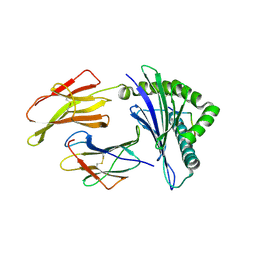 | | MHC A02 Allele presenting MTSAIGILPV | | 分子名称: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-2 alpha chain, ... | | 著者 | Rizkallah, P.J, Sewell, A.K. | | 登録日 | 2018-03-26 | | 公開日 | 2019-09-04 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Peptide Super-Agonist Enhances T-Cell Responses to Melanoma.
Front Immunol, 10, 2019
|
|
7TIX
 
 | | Crystal structure of SARS-CoV-2 3CL in complex with inhibitor EB56 | | 分子名称: | 3C-like proteinase nsp5, MAGNESIUM ION, N~2~-{[(naphthalen-2-yl)methoxy]carbonyl}-N-{(2S)-1-oxo-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | | 著者 | Forouhar, F, Liu, H, Iketani, S, Zack, A, Khanizeman, N, Bednarova, E, Fowler, B, Hong, S.J, Mohri, H, Nair, M.S, Huang, Y, Tay, N.E.S, Lee, S, Karan, C, Resnick, S.J, Quinn, C, Li, W, Shion, H, Jurtschenko, C, Lauber, M.A, McDonald, T, Stokes, M.E, Hurst, B, Rovis, T, Chavez, A, Ho, D.D, Stockwell, B.R. | | 登録日 | 2022-01-14 | | 公開日 | 2022-05-04 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Development of optimized drug-like small molecule inhibitors of the SARS-CoV-2 3CL protease for treatment of COVID-19.
Nat Commun, 13, 2022
|
|
6G44
 
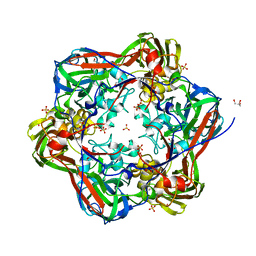 | | Crystal structure of mavirus major capsid protein lacking the C-terminal domain | | 分子名称: | GLYCEROL, Putative major capsid protein, SULFATE ION | | 著者 | Born, D, Reuter, L, Meinhart, A, Reinstein, J. | | 登録日 | 2018-03-26 | | 公開日 | 2018-07-04 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Capsid protein structure, self-assembly, and processing reveal morphogenesis of the marine virophage mavirus.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6NIW
 
 | | Crystal structure of P[6] rotavirus | | 分子名称: | DI(HYDROXYETHYL)ETHER, Protease-sensitive outer capsid protein | | 著者 | Xu, S, Liu, Y, Lakamp, L, Ahmed, L, Jiang, X, Kennedy, M.A. | | 登録日 | 2019-01-01 | | 公開日 | 2020-01-08 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Molecular basis of P[II] major human rotavirus VP8* domain recognition of histo-blood group antigens.
Plos Pathog., 16, 2020
|
|
5KF2
 
 | | X-ray structure of a glucosamine N-Acetyltransferase from Clostridium acetobutylicum, apo form, pH 8 | | 分子名称: | 1,2-ETHANEDIOL, ACETYL COENZYME *A, COENZYME A, ... | | 著者 | Holden, H.M, Thoden, J.B, Dopkins, B.J, Tipton, P.A. | | 登録日 | 2016-06-11 | | 公開日 | 2016-07-06 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural Studies on a Glucosamine/Glucosaminide N-Acetyltransferase.
Biochemistry, 55, 2016
|
|
7AN9
 
 | |
6NX4
 
 | | Structure of the C-terminal Helical Repeat Domain of Eukaryotic Elongation Factor 2 Kinase (eEF-2K) | | 分子名称: | Eukaryotic elongation factor 2 kinase | | 著者 | Piserchio, A, Will, N, Giles, D.H, Hajredini, F, Dalby, K.N, Ghose, R. | | 登録日 | 2019-02-08 | | 公開日 | 2019-05-29 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution Structure of the Carboxy-Terminal Tandem Repeat Domain of Eukaryotic Elongation Factor 2 Kinase and Its Role in Substrate Recognition.
J.Mol.Biol., 431, 2019
|
|
5KGI
 
 | |
7ALE
 
 | | Crystal structure of human PAICS in complex with inhibitor 69 | | 分子名称: | (2~{S})-2-[[5-azanyl-1-[(2~{R},3~{R},4~{S},5~{R})-3,4-bis(oxidanyl)-5-(phosphonooxymethyl)oxolan-2-yl]imidazol-4-yl]car bonylamino]butanedioic acid, 2-azanyl-~{N}-[2-bromanyl-5-[4-[3-(dimethylamino)propylsulfonyl]piperazin-1-yl]phenyl]-1,3-oxazole-4-carboxamide, Multifunctional protein ADE2 | | 著者 | Skerlova, J, Marttila, P, Unterlass, J, Jemth, A.-S, Henriksson, M, Wakchaure, P, Grube, M, Warpman Berglund, U, Homan, E, Helleday, T, Stenmark, P. | | 登録日 | 2020-10-06 | | 公開日 | 2022-04-20 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.95 Å) | | 主引用文献 | Cellular and biochemical validation of a potent PAICS inhibitor
To Be Published
|
|
