1CMR
 
 | | NMR SOLUTION STRUCTURE OF A CHIMERIC PROTEIN, DESIGNED BY TRANSFERRING A FUNCTIONAL SNAKE BETA-HAIRPIN INTO A SCORPION ALPHA/BETA SCAFFOLD (PH 3.5, 20C), NMR, 18 STRUCTURES | | 分子名称: | CHARYBDOTOXIN, ALPHA CHIMERA | | 著者 | Zinn-Justin, S, Guenneugues, M, Drakopoulou, E, Gilquin, B, Vita, C, Menez, A. | | 登録日 | 1996-03-15 | | 公開日 | 1996-08-01 | | 最終更新日 | 2021-11-03 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Transfer of a beta-hairpin from the functional site of snake curaremimetic toxins to the alpha/beta scaffold of scorpion toxins: three-dimensional solution structure of the chimeric protein.
Biochemistry, 35, 1996
|
|
7G17
 
 | | Crystal Structure of human FABP4 in complex with 2-(1H-indol-7-yloxymethyl)-1,3-thiazole-4-carboxylic acid | | 分子名称: | 1,2-ETHANEDIOL, 2-{[(1H-indol-7-yl)oxy]methyl}-1,3-thiazole-4-carboxylic acid, Fatty acid-binding protein, ... | | 著者 | Ehler, A, Benz, J, Obst, U, Brunner, M, Rudolph, M.G. | | 登録日 | 2023-04-27 | | 公開日 | 2023-06-14 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.05 Å) | | 主引用文献 | Crystal Structure of a human FABP4 complex
To be published
|
|
7ZQW
 
 | | Structure of the SARS-CoV-1 main protease in complex with AG7404 | | 分子名称: | 3C-like proteinase nsp5, ethyl (4R)-4-({(2S)-2-[3-{[(5-methyl-1,2-oxazol-3-yl)carbonyl]amino}-2-oxopyridin-1(2H)-yl]pent-4-ynoyl}amino)-5-[(3S)-2-oxopyrrolidin-3-yl]pentanoate | | 著者 | Muriel-Goni, S, Fabrega-Ferrer, M, Herrera-Morande, A, Coll, M. | | 登録日 | 2022-05-03 | | 公開日 | 2022-12-28 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (2.53 Å) | | 主引用文献 | Structure and inhibition of SARS-CoV-1 and SARS-CoV-2 main proteases by oral antiviral compound AG7404.
Antiviral Res., 208, 2022
|
|
6GWM
 
 | | Solution structure of rat RIP2 caspase recruitment domain | | 分子名称: | Receptor-interacting serine/threonine-protein kinase 2 | | 著者 | Mineev, K.S, Goncharuk, S.A, Artemieva, L.E, Arseniev, A.S. | | 登録日 | 2018-06-25 | | 公開日 | 2018-10-31 | | 最終更新日 | 2024-06-19 | | 実験手法 | SOLUTION NMR | | 主引用文献 | CARD domain of rat RIP2 kinase: Refolding, solution structure, pH-dependent behavior and protein-protein interactions.
PLoS ONE, 13, 2018
|
|
7G0I
 
 | | Crystal Structure of human FABP4 in complex with 5-phenoxy-6-(2,2,2-trifluoroethoxy)-2-(trifluoromethyl)pyridine-3-carboxylic acid | | 分子名称: | 5-phenoxy-6-(2,2,2-trifluoroethoxy)-2-(trifluoromethyl)pyridine-3-carboxylic acid, DIMETHYL SULFOXIDE, Fatty acid-binding protein, ... | | 著者 | Ehler, A, Benz, J, Obst, U, Richter, H, Rudolph, M.G. | | 登録日 | 2023-04-27 | | 公開日 | 2023-06-14 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.07 Å) | | 主引用文献 | Crystal Structure of a human FABP4 complex
To be published
|
|
6ZCH
 
 | |
5EOW
 
 | | Crystal Structure of 6-Hydroxynicotinic Acid 3-Monooxygenase from Pseudomonas putida KT2440 | | 分子名称: | 6-hydroxynicotinate 3-monooxygenase, FLAVIN-ADENINE DINUCLEOTIDE | | 著者 | Yuen, M.E, Zhen, W, Gerwig, T.J, Story, R.W, Kopp, M, Nakamoto, K, Snider, M.J, Hicks, K.A. | | 登録日 | 2015-11-10 | | 公開日 | 2016-06-08 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural and Biochemical Characterization of 6-Hydroxynicotinic Acid 3-Monooxygenase, A Novel Decarboxylative Hydroxylase Involved in Aerobic Nicotinate Degradation.
Biochemistry, 55, 2016
|
|
8PFW
 
 | |
7G0L
 
 | | Crystal Structure of human FABP4 in complex with 3-[(4-methyl-3-phenoxyphenyl)methyl]-1H-pyrazol-5-ol | | 分子名称: | 3-[(4-methyl-3-phenoxyphenyl)methyl]-1H-pyrazol-5-ol, FORMIC ACID, Fatty acid-binding protein, ... | | 著者 | Ehler, A, Benz, J, Obst, U, Sweeney, Z, Rudolph, M.G. | | 登録日 | 2023-04-27 | | 公開日 | 2023-06-14 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.18 Å) | | 主引用文献 | Crystal Structure of a human FABP4 complex
To be published
|
|
1CYF
 
 | |
7G0T
 
 | | Crystal Structure of human FABP4 in complex with 1-[[4-chloro-2-(trifluoromethyl)phenyl]carbamoylamino]cyclopentane-1-carboxylic acid | | 分子名称: | 1-{[4-chloro-2-(trifluoromethyl)phenyl]carbamamido}cyclopentane-1-carboxylic acid, DIMETHYL SULFOXIDE, FORMIC ACID, ... | | 著者 | Ehler, A, Benz, J, Obst, U, Maurer, M, Rudolph, M.G. | | 登録日 | 2023-04-27 | | 公開日 | 2023-06-14 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Crystal Structure of a human FABP4 complex
To be published
|
|
7G1A
 
 | | Crystal Structure of human FABP4 in complex with 2-[(3S)-oxolan-3-yl]oxy-4-phenyl-3-(1H-tetrazol-5-yl)-6,7,8,9-tetrahydro-5H-cyclohepta[b]pyridine | | 分子名称: | (3M)-2-{[(3S)-oxolan-3-yl]oxy}-4-phenyl-3-(1H-tetrazol-5-yl)-6,7,8,9-tetrahydro-5H-cyclohepta[b]pyridine, DIMETHYL SULFOXIDE, Fatty acid-binding protein, ... | | 著者 | Ehler, A, Benz, J, Obst, U, Buettelmann, B, Rudolph, M.G. | | 登録日 | 2023-04-27 | | 公開日 | 2023-06-14 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.12 Å) | | 主引用文献 | Crystal Structure of a human FABP4 complex
To be published
|
|
7LP3
 
 | | Structure of Nedd4L WW3 domain | | 分子名称: | Angiomotin, E3 ubiquitin-protein ligase NEDD4-like, SULFATE ION | | 著者 | Alian, A, Alam, S.L, Thompson, T, Rheinemann, L, Sundquist, W.I. | | 登録日 | 2021-02-11 | | 公開日 | 2021-07-28 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.61 Å) | | 主引用文献 | Interactions between AMOT PPxY motifs and NEDD4L WW domains function in HIV-1 release.
J.Biol.Chem., 297, 2021
|
|
8VWS
 
 | |
7G1L
 
 | | Crystal Structure of human FABP4 in complex with 6-(1,3-benzodioxol-5-ylmethyl)-3-sulfanyl-1,2,4-triazin-5-ol | | 分子名称: | 6-[(2H-1,3-benzodioxol-5-yl)methyl]-3-sulfanyl-1,2,4-triazin-5-ol, FORMIC ACID, Fatty acid-binding protein, ... | | 著者 | Ehler, A, Benz, J, Obst, U, Rudolph, M.G. | | 登録日 | 2023-04-27 | | 公開日 | 2023-06-14 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (0.98 Å) | | 主引用文献 | Crystal Structure of a human FABP4 complex
To be published
|
|
7LP2
 
 | | Structure of Nedd4L WW3 domain | | 分子名称: | Angiomotin, E3 ubiquitin-protein ligase, GLYCEROL, ... | | 著者 | Alian, A, Alam, S.L, Thompson, T, Rheinemann, L, Sundquist, W.I. | | 登録日 | 2021-02-11 | | 公開日 | 2021-07-28 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.88 Å) | | 主引用文献 | Interactions between AMOT PPxY motifs and NEDD4L WW domains function in HIV-1 release.
J.Biol.Chem., 297, 2021
|
|
7TJ1
 
 | | Crystal Structure of the Putative Fluoride Ion Transporter CrcB Bab1_1389 from Brucella abortus | | 分子名称: | 1,2-ETHANEDIOL, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Kim, Y, Tesar, C, Pastore, T, Endres, M, Babnigg, G, Crosson, S, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2022-01-14 | | 公開日 | 2022-01-26 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal Structure of the Putative Fluoride Ion Transporter CrcB Bab1_1389 from Brucella abortus
To Be Published
|
|
5EHF
 
 | | Laccase from Antrodiella faginea | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, ... | | 著者 | Polyakov, K.M, Glazunova, O.A, Fedorova, T.V, Dorovatovskii, P.V, Koroleva, O.V. | | 登録日 | 2015-10-28 | | 公開日 | 2015-12-23 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Structure-function study of two new middle-redox potential laccases from basidiomycetes Antrodiella faginea and Steccherinum murashkinskyi.
Int. J. Biol. Macromol., 118, 2018
|
|
7G1B
 
 | | Crystal Structure of human FABP4 in complex with 2-[1-[(3-chlorophenyl)methyl]indol-2-yl]cyclopropane-1-carboxylic acid | | 分子名称: | (1S,2S)-2-{1-[(3-chlorophenyl)methyl]-1H-indol-2-yl}cyclopropane-1-carboxylic acid, Fatty acid-binding protein, adipocyte | | 著者 | Ehler, A, Benz, J, Obst, U, Rudolph, M.G. | | 登録日 | 2023-04-27 | | 公開日 | 2023-06-14 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.17 Å) | | 主引用文献 | Crystal Structure of a human FABP4 complex
To be published
|
|
7G0K
 
 | | Crystal Structure of human FABP4 in complex with (2R)-1-[(3,5-dichloro-2-phenylphenyl)carbamoyl]pyrrolidine-2-carboxylic acid | | 分子名称: | 1-[(4,6-dichloro[1,1'-biphenyl]-2-yl)carbamoyl]-D-proline, FORMIC ACID, Fatty acid-binding protein, ... | | 著者 | Ehler, A, Benz, J, Obst, U, Obst-Sander, U, Rudolph, M.G. | | 登録日 | 2023-04-27 | | 公開日 | 2023-06-14 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.13 Å) | | 主引用文献 | Crystal Structure of a human FABP4 complex
To be published
|
|
7FXM
 
 | | Crystal Structure of human FABP4 in complex with 5-cyclohexyl-2-hydroxybenzoic acid | | 分子名称: | 5-cyclohexyl-2-hydroxybenzoate, Fatty acid-binding protein, adipocyte, ... | | 著者 | Ehler, A, Benz, J, Obst, U, Urban, R, Rudolph, M.G. | | 登録日 | 2023-04-27 | | 公開日 | 2023-06-14 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.19 Å) | | 主引用文献 | Crystal Structure of a human FABP4 complex
To be published
|
|
8PM2
 
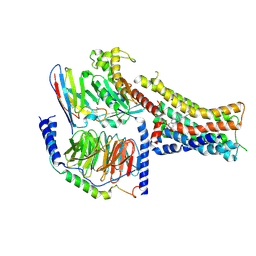 | | Structure of the murine trace amine-associated receptor TAAR7f bound to N,N-dimethylcyclohexylamine (DMCH) in complex with mini-Gs trimeric G protein | | 分子名称: | CHOLESTEROL HEMISUCCINATE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Gusach, A, Lee, Y, Edwards, P.C, Huang, F, Weyand, S.N, Tate, C.G. | | 登録日 | 2023-06-28 | | 公開日 | 2023-08-09 | | 最終更新日 | 2024-10-23 | | 実験手法 | ELECTRON MICROSCOPY (2.92 Å) | | 主引用文献 | Molecular recognition of an aversive odorant by the murine trace amine-associated receptor TAAR7f.
Biorxiv, 2023
|
|
7FVV
 
 | | Crystal Structure of human FABP4 in complex with (1S,2R)-2-[(5-carbamoyl-3-ethoxycarbonyl-4-methyl-2-thienyl)carbamoyl]cyclohexanecarboxylic acid | | 分子名称: | (1R,2S)-2-{[5-carbamoyl-3-(ethoxycarbonyl)-4-methylthiophen-2-yl]carbamoyl}cyclohexane-1-carboxylic acid, Fatty acid-binding protein, adipocyte | | 著者 | Ehler, A, Benz, J, Obst, U, Rudolph, M.G. | | 登録日 | 2023-04-27 | | 公開日 | 2023-06-14 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.08 Å) | | 主引用文献 | Crystal Structure of a human FABP4 complex
To be published
|
|
7FW6
 
 | | Crystal Structure of human FABP4 in complex with 2-[(3-chlorophenoxy)methyl]-4-phenoxycyclohexane-1-carboxylic acid | | 分子名称: | (1R,2S,4S)-2-[(3-chlorophenoxy)methyl]-4-phenoxycyclohexane-1-carboxylic acid, Fatty acid-binding protein, adipocyte, ... | | 著者 | Ehler, A, Benz, J, Obst, U, Buettelmann, B, Rudolph, M.G. | | 登録日 | 2023-04-27 | | 公開日 | 2023-06-14 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.04 Å) | | 主引用文献 | Crystal Structure of a human FABP4 complex
To be published
|
|
7Q37
 
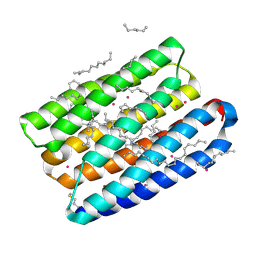 | | Crystal structure of proton pump MAR rhodopsin pressurized with krypton | | 分子名称: | Bacteriorhodopsin, EICOSANE, HEXANE, ... | | 著者 | Melnikov, I, Rulev, M, Astashkin, R, Kovalev, K, Carpentier, P, Gordeliy, V, Popov, A. | | 登録日 | 2021-10-27 | | 公開日 | 2022-04-27 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | High-pressure crystallography shows noble gas intervention into protein-lipid interaction and suggests a model for anaesthetic action.
Commun Biol, 5, 2022
|
|
