6CUZ
 
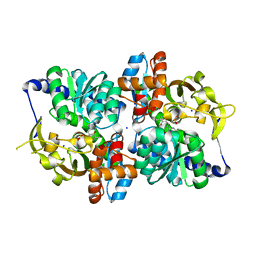 | | Engineered TrpB from Pyrococcus furiosus, PfTrpB7E6 with (2S,3R)-ethylserine bound as the amino-acrylate | | 分子名称: | (2E)-2-[(E)-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)amino]pent-2-enoic acid, PHOSPHATE ION, SODIUM ION, ... | | 著者 | Scheele, R.A, Buller, A.R, Boville, C.E, Arnold, F.H. | | 登録日 | 2018-03-27 | | 公開日 | 2018-09-26 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Engineered Biosynthesis of beta-Alkyl Tryptophan Analogues.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
5D5L
 
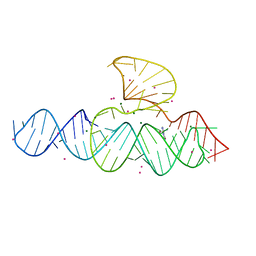 | |
6CDD
 
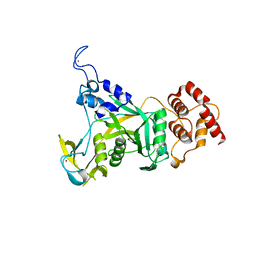 | |
5UQ3
 
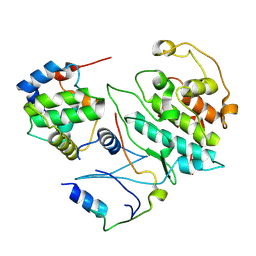 | |
5D8H
 
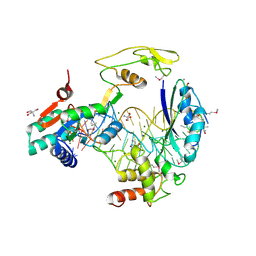 | |
4UWU
 
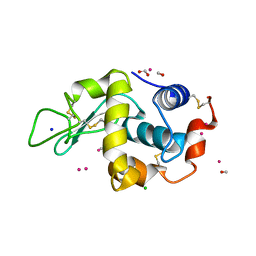 | | Lysozyme soaked with a ruthenium based CORM with a pyridine ligand (complex 7) | | 分子名称: | CARBON MONOXIDE, CHLORIDE ION, FORMIC ACID, ... | | 著者 | Santos, M.F.A, Mukhopadhyay, A, Romao, M.J, Romao, C.C, Santos-Silva, T. | | 登録日 | 2014-08-14 | | 公開日 | 2014-12-10 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | A Contribution to the Rational Design of Ru(Co)3Cl2L Complexes for in Vivo Delivery of Co.
Dalton Trans, 44, 2015
|
|
5UXC
 
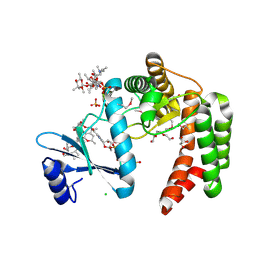 | | Crystal structure of macrolide 2'-phosphotransferase MphH from Brachybacterium faecium in complex with GDP | | 分子名称: | AZITHROMYCIN, CHLORIDE ION, GLYCEROL, ... | | 著者 | Stogios, P.J, Skarina, T, Wawrzak, Z, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2017-02-22 | | 公開日 | 2017-08-16 | | 最終更新日 | 2019-12-11 | | 実験手法 | X-RAY DIFFRACTION (1.72 Å) | | 主引用文献 | The evolution of substrate discrimination in macrolide antibiotic resistance enzymes.
Nat Commun, 9, 2018
|
|
5V86
 
 | | Structure of DCN1 bound to NAcM-OPT | | 分子名称: | Lysozyme,DCN1-like protein 1, N-benzyl-N-(1-butylpiperidin-4-yl)-N'-(3,4-dichlorophenyl)urea | | 著者 | Guy, R.K, Schulman, B.A, Scott, D.C, Hammill, J.T. | | 登録日 | 2017-03-21 | | 公開日 | 2017-05-24 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.374 Å) | | 主引用文献 | Blocking an N-terminal acetylation-dependent protein interaction inhibits an E3 ligase.
Nat. Chem. Biol., 13, 2017
|
|
6CIG
 
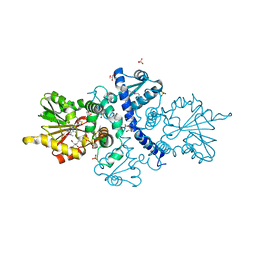 | | CRYSTAL STRUCTURE ANALYSIS OF SELENOMETHIONINE SUBSTITUTED ISOFLAVONE O-METHYLTRANSFERASE | | 分子名称: | GLYCEROL, Isoflavone-7-O-methyltransferase 8, N-(TRIS(HYDROXYMETHYL)METHYL)-3-AMINOPROPANESULFONIC ACID, ... | | 著者 | Zubieta, C, Dixon, R.A, Shabalin, I.G, Kowiel, M, Porebski, P.J, Noel, J.P. | | 登録日 | 2018-02-23 | | 公開日 | 2018-03-07 | | 最終更新日 | 2022-03-23 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Structures of two natural product methyltransferases reveal the basis for substrate specificity in plant O-methyltransferases.
Nat. Struct. Biol., 8, 2001
|
|
6CF6
 
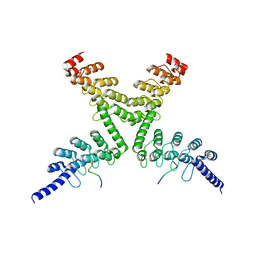 | | RNF146 TBM-Tankyrase ARC2-3 complex | | 分子名称: | RNF146, Tankyrase-1 | | 著者 | Da Rosa, P.A, Xu, W. | | 登録日 | 2018-02-13 | | 公開日 | 2018-04-18 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | Structural basis for tankyrase-RNF146 interaction reveals noncanonical tankyrase-binding motifs.
Protein Sci., 27, 2018
|
|
6CP7
 
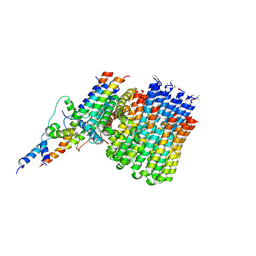 | | Monomer yeast ATP synthase Fo reconstituted in nanodisc generated from masked refinement. | | 分子名称: | ATP synthase protein 8, ATP synthase subunit 4, mitochondrial, ... | | 著者 | Srivastava, A.P, Luo, M, Symersky, J, Liao, M.F, Mueller, D.M. | | 登録日 | 2018-03-13 | | 公開日 | 2018-04-11 | | 最終更新日 | 2020-01-15 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | High-resolution cryo-EM analysis of the yeast ATP synthase in a lipid membrane.
Science, 360, 2018
|
|
6CQM
 
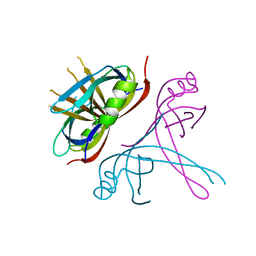 | | Crystal Structure of mitochondrial single-stranded DNA binding proteins from S. cerevisiae, Rim1 (Form2) | | 分子名称: | Single-stranded DNA-binding protein RIM1, mitochondrial | | 著者 | Singh, S.P, Kukshal, V, Bona, P.D, Lytle, A.K, Edwin, A, Galletto, R. | | 登録日 | 2018-03-15 | | 公開日 | 2018-05-30 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | The mitochondrial single-stranded DNA binding protein from S. cerevisiae, Rim1, does not form stable homo-tetramers and binds DNA as a dimer of dimers.
Nucleic Acids Res., 46, 2018
|
|
6CRM
 
 | |
6CRZ
 
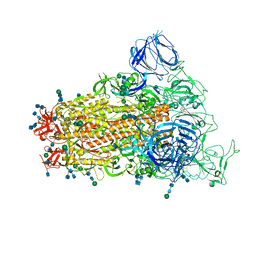 | | SARS Spike Glycoprotein, Trypsin-cleaved, Stabilized variant, C3 symmetry | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Fibritin, ... | | 著者 | Kirchdoerfer, R.N, Wang, N, Pallesen, J, Turner, H.L, Cottrell, C.A, McLellan, J.S, Ward, A.B. | | 登録日 | 2018-03-19 | | 公開日 | 2018-04-11 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Stabilized coronavirus spikes are resistant to conformational changes induced by receptor recognition or proteolysis.
Sci Rep, 8, 2018
|
|
5UNF
 
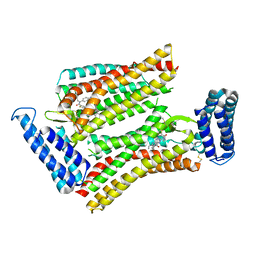 | | XFEL structure of human angiotensin II type 2 receptor (Monoclinic form) in complex with compound 1 (N-benzyl-N-(2-ethyl-4-oxo-3-{[2'-(2H-tetrazol-5-yl)[1,1'-biphenyl]-4-yl]) | | 分子名称: | Chimera protein of Type-2 angiotensin II receptor and Soluble cytochrome b562, N-benzyl-N-(2-ethyl-4-oxo-3-{[2'-(2H-tetrazol-5-yl)[1,1'-biphenyl]-4-yl]methyl}-3,4-dihydroquinazolin-6-yl)thiophene-2-carboxamide | | 著者 | Zhang, H, Han, G.W, Batyuk, A, Ishchenko, A, White, K.L, Patel, N, Sadybekov, A, Zamlynny, B, Rudd, M.T, Hollenstein, K, Tolstikova, A, White, T.A, Hunter, M.S, Weierstall, U, Liu, W, Babaoglu, K, Moore, E.L, Katz, R.D, Shipman, J.M, Garcia-Calvo, M, Sharma, S, Sheth, P, Soisson, S.M, Stevens, R.C, Katritch, V, Cherezov, V. | | 登録日 | 2017-01-30 | | 公開日 | 2017-04-05 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural basis for selectivity and diversity in angiotensin II receptors.
Nature, 544, 2017
|
|
5VB8
 
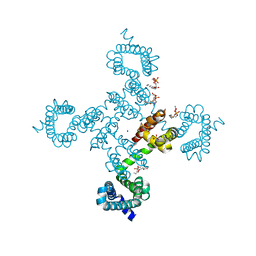 | |
4V2T
 
 | | Membrane embedded pleurotolysin pore with 13 fold symmetry | | 分子名称: | PLEUROTOLYSIN A, PLEUROTOLYSIN B | | 著者 | Lukoyanova, N, Kondos, S.C, Farabella, I, Law, R.H.P, Reboul, C.F, Caradoc-Davies, T.T, Spicer, B.A, Kleifeld, O, Perugini, M, Ekkel, S, Hatfaludi, T, Oliver, K, Hotze, E.M, Tweten, R.K, Whisstock, J.C, Topf, M, Dunstone, M.A, Saibil, H.R. | | 登録日 | 2014-10-15 | | 公開日 | 2015-02-18 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (11 Å) | | 主引用文献 | Conformational Changes During Pore Formation by the Perforin-Related Protein Pleurotolysin.
Plos Biol., 13, 2015
|
|
4V4R
 
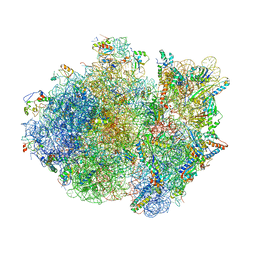 | | Crystal structure of the whole ribosomal complex. | | 分子名称: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Petry, S, Brodersen, D.E, Murphy IV, F.V, Dunham, C.M, Selmer, M, Tarry, M.J, Kelley, A.C, Ramakrishnan, V. | | 登録日 | 2005-09-30 | | 公開日 | 2014-07-09 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (5.9 Å) | | 主引用文献 | Crystal Structures of the Ribosome in Complex with Release Factors RF1 and RF2 Bound to a Cognate Stop Codon.
Cell(Cambridge,Mass.), 123, 2005
|
|
6CTA
 
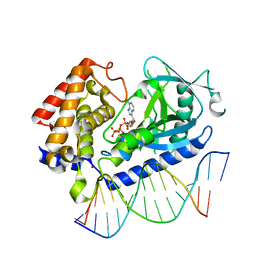 | | Structure of the human cGAS-DNA complex with ATP | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Cyclic GMP-AMP synthase, DNA (5'-D(*AP*AP*AP*TP*TP*GP*CP*CP*GP*AP*AP*GP*AP*CP*GP*A)-3'), ... | | 著者 | Zhou, W, Whiteley, A.T, de Oliveira Mann, C.C, Morehouse, B.R, Mekalanos, J.J, Kranzusch, P.J. | | 登録日 | 2018-03-22 | | 公開日 | 2018-07-18 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.779 Å) | | 主引用文献 | Structure of the Human cGAS-DNA Complex Reveals Enhanced Control of Immune Surveillance.
Cell, 174, 2018
|
|
4UZV
 
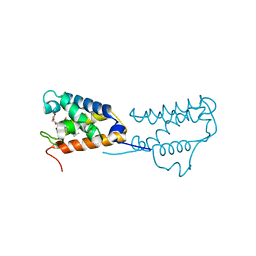 | | Structure of a triple mutant of ASV-TfTrHb | | 分子名称: | ACETATE ION, HEMOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Baiocco, P, Bonamore, A, Sciamanna, N, Ilari, A, Boechi, L, Boffi, A, Smulevich, G, Feis, A. | | 登録日 | 2014-09-09 | | 公開日 | 2014-09-17 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (3.4 Å) | | 主引用文献 | Role of Local Structure and Dynamics of Small Ligand Migration in Proteins: A Study of a Mutated Truncated Hemoprotein from Thermobifida Fusca by Time Resolved Mir Spectroscopy.
J.Phys.Chem.B, 118, 2014
|
|
5V1M
 
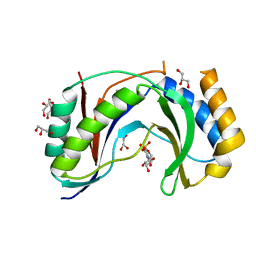 | | Structure of human Usb1 with uridine 5'-monophosphate | | 分子名称: | CHLORIDE ION, GLYCEROL, U6 snRNA phosphodiesterase, ... | | 著者 | Montemayor, E.J, Didychuk, A.L, Butcher, S.E. | | 登録日 | 2017-03-02 | | 公開日 | 2017-08-16 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.47 Å) | | 主引用文献 | Usb1 controls U6 snRNP assembly through evolutionarily divergent cyclic phosphodiesterase activities.
Nat Commun, 8, 2017
|
|
4V4S
 
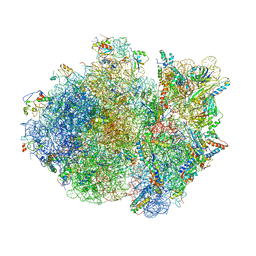 | | Crystal structure of the whole ribosomal complex. | | 分子名称: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Petry, S, Brodersen, D.E, Murphy IV, F.V, Dunham, C.M, Selmer, M, Tarry, M.J, Kelley, A.C, Ramakrishnan, V. | | 登録日 | 2005-10-12 | | 公開日 | 2014-07-09 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (6.76 Å) | | 主引用文献 | Crystal Structures of the Ribosome in Complex with Release Factors RF1 and RF2 Bound to a Cognate Stop Codon.
Cell(Cambridge,Mass.), 123, 2005
|
|
128D
 
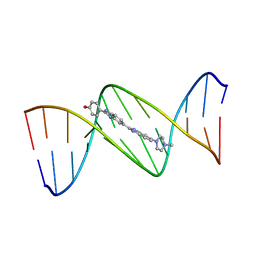 | | MOLECULAR STRUCTURE OF D(CGC[E6G]AATTCGCG) COMPLEXED WITH HOECHST 33258 | | 分子名称: | 2'-(4-HYDROXYPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, DNA (5'-D(*CP*GP*CP*(G36)P*AP*AP*TP*TP*CP*GP*CP*G)-3') | | 著者 | Sriram, M, Van Der Marel, G.A, Roelen, H.L.P.F, Van Boom, J.H, Wang, A.H.-J. | | 登録日 | 1993-06-30 | | 公開日 | 1994-01-15 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Conformation of B-DNA containing O6-ethyl-G-C base pairs stabilized by minor groove binding drugs: molecular structure of d(CGC[e6G]AATTCGCG complexed with Hoechst 33258 or Hoechst 33342.
EMBO J., 11, 1992
|
|
129D
 
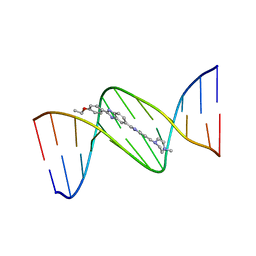 | | DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3') COMPLEXED WITH HOECHST 33342 | | 分子名称: | 2'-(4-ETHOXYPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3') | | 著者 | Sriram, M, Van Der Marel, G.A, Roelen, H.L.P.F, Van Boom, J.H, Wang, A.H.-J. | | 登録日 | 1993-06-30 | | 公開日 | 1994-01-15 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Conformation of B-DNA containing O6-ethyl-G-C base pairs stabilized by minor groove binding drugs: molecular structure of d(CGC[e6G]AATTCGCG complexed with Hoechst 33258 or Hoechst 33342.
EMBO J., 11, 1992
|
|
4V62
 
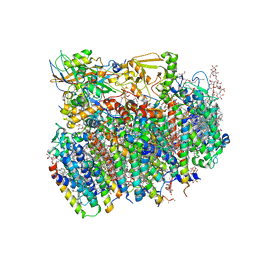 | | Crystal Structure of cyanobacterial Photosystem II | | 分子名称: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | 著者 | Guskov, A, Gabdulkhakov, A, Kern, J, Broser, M, Zouni, A, Saenger, W. | | 登録日 | 2008-01-17 | | 公開日 | 2014-07-09 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Cyanobacterial photosystem II at 2.9-A resolution and the role of quinones, lipids, channels and chloride
Nat.Struct.Mol.Biol., 16, 2009
|
|
