6X1Q
 
 | | 1.8 Angstrom resolution structure of b-galactosidase with a 200 kV cryoARM electron microscope | | Descriptor: | Beta-galactosidase, MAGNESIUM ION, SODIUM ION | | Authors: | Merk, A, Fukumura, T, Zhu, X, Darling, J, Grisshammer, R, Ognjenovic, J, Subramaniam, S. | | Deposit date: | 2020-05-19 | | Release date: | 2020-07-01 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (1.8 Å) | | Cite: | 1.8 angstrom resolution structure of beta-galactosidase with a 200 kV CRYO ARM electron microscope.
Iucrj, 7, 2020
|
|
7T69
 
 | | Crystal structure of Avr3 (SIX1) from Fusarium oxysporum f. sp. lycopersici | | Descriptor: | Avr3 (SIX1), Secreted in xylem 1, SULFATE ION | | Authors: | Yu, D.S, Outram, M.A, Ericsson, D.J, Jones, D.A, Williams, S.J. | | Deposit date: | 2021-12-13 | | Release date: | 2023-01-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | The structural repertoire of Fusarium oxysporum f. sp. lycopersici effectors revealed by experimental and computational studies
Elife, 2023
|
|
4YUO
 
 | | High-resolution multiconformer synchrotron model of CypA at 273 K | | Descriptor: | Peptidyl-prolyl cis-trans isomerase A | | Authors: | Keedy, D.A, Kenner, L.R, Warkentin, M, Woldeyes, R.A, Thompson, M.C, Brewster, A.S, Van Benschoten, A.H, Baxter, E.L, Hopkins, J.B, Uervirojnangkoorn, M, McPhillips, S.E, Song, J, Mori, R.A, Holton, J.M, Weis, W.I, Brunger, A.T, Soltis, M, Lemke, H, Gonzalez, A, Sauter, N.K, Cohen, A.E, van den Bedem, H, Thorne, R.E, Fraser, J.S. | | Deposit date: | 2015-03-18 | | Release date: | 2015-05-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Mapping the conformational landscape of a dynamic enzyme by multitemperature and XFEL crystallography.
Elife, 4, 2015
|
|
8CXM
 
 | | Cryo-EM structure of the supercoiled E. coli K12 flagellar filament core, Normal waveform | | Descriptor: | Flagellin | | Authors: | Sonani, R.R, Kreutzberger, M.A.B, Sebastian, A.L, Scharf, B, Egelman, E.H. | | Deposit date: | 2022-05-21 | | Release date: | 2022-09-07 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Convergent evolution in the supercoiling of prokaryotic flagellar filaments.
Cell, 185, 2022
|
|
7QAA
 
 | | Crystal structure of RARalpha/RXRalpha ligand binding domain heterodimer in complex with BMS614 and oleic acid | | Descriptor: | 4-[(4,4-DIMETHYL-1,2,3,4-TETRAHYDRO-[1,2']BINAPTHALENYL-7-CARBONYL)-AMINO]-BENZOIC ACID, Isoform Alpha-1-deltaBC of Retinoic acid receptor alpha, OLEIC ACID, ... | | Authors: | le Maire, A, Vivat, V, Guee, L, Blanc, P, Malosse, C, Chamot-Rooke, J, Germain, P, Bourguet, w. | | Deposit date: | 2021-11-16 | | Release date: | 2022-10-05 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Design and in vitro characterization of RXR variants as tools to investigate the biological role of endogenous rexinoids.
J.Mol.Endocrinol., 69, 2022
|
|
7T6A
 
 | | Crystal structure of Avr1 (SIX4) from Fusarium oxysporum f. sp. lycopersici | | Descriptor: | Avr1 (FolSIX4), Avirulence protein 1, Avr1 (SIX4), ... | | Authors: | Yu, D.S, Outram, M.A, Ericsson, D.J, Jones, D.A, Williams, S.J. | | Deposit date: | 2021-12-13 | | Release date: | 2023-01-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The structural repertoire of Fusarium oxysporum f. sp. lycopersici effectors revealed by experimental and computational studies
Elife, 2023
|
|
8U0P
 
 | | Synaptic complex of human DNA polymerase Lambda DL variant engaged on a noncomplementary DNA double-strand break | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, CHLORIDE ION, DNA (5'-D(*CP*AP*GP*TP*AP*C)-3'), ... | | Authors: | Kaminski, A.M, Pedersen, L.C, Bebenek, K, Kunkel, T.A, Chiruvella, K.K, Ramsden, D.A. | | Deposit date: | 2023-08-29 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | DNA polymerase lambda Loop1 variant yields unexpected gain-of-function capabilities in nonhomologous end-joining.
DNA Repair (Amst), 136, 2024
|
|
6V5L
 
 | | The HADDOCK structure model of GDP KRas in complex with its allosteric inhibitor E22 | | Descriptor: | (2R)-2-[2-(1H-indole-3-carbonyl)hydrazinyl]-2-phenylacetamide, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Wang, X, Gupta, A.K, Prakash, P, Putkey, J.P, Gorfe, A.A. | | Deposit date: | 2019-12-04 | | Release date: | 2019-12-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Multi target ensemble based virtual screening yields novel allosteric KRAS inhibitors at high success rate
Chemical Biology & Drug Design, 94, 2019
|
|
5JJS
 
 | | Dengue 3 NS5 protein with compound 27 | | Descriptor: | 1,2-ETHANEDIOL, 5-[5-(3-hydroxyprop-1-yn-1-yl)thiophen-2-yl]-2,4-dimethoxy-N-{[(1R,3R)-3-methoxycyclohexyl]sulfonyl}benzamide, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Lescar, J, El Sahili, A. | | Deposit date: | 2016-04-25 | | Release date: | 2016-07-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Potent Allosteric Dengue Virus NS5 Polymerase Inhibitors: Mechanism of Action and Resistance Profiling
Plos Pathog., 12, 2016
|
|
6X4E
 
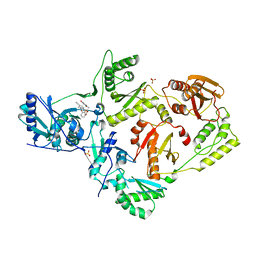 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with methyl 2-(6-cyano-3-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)-4-methylnaphthalen-1-yl)acetate (JLJ681), a Non-nucleoside Inhibitor | | Descriptor: | Reverse transcriptase/ribonuclease H, SULFATE ION, methyl (6-cyano-3-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]phenoxy}-4-methylnaphthalen-1-yl)acetate, ... | | Authors: | Chan, A.H, Duong, V.N, Ippolito, J.A, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2020-05-22 | | Release date: | 2020-07-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural investigation of 2-naphthyl phenyl ether inhibitors bound to WT and Y181C reverse transcriptase highlights key features of the NNRTI binding site.
Protein Sci., 29, 2020
|
|
5MAK
 
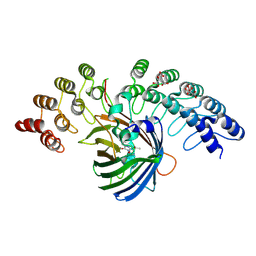 | | GFP-binding DARPin fusion gc_R7 | | Descriptor: | CITRIC ACID, Green fluorescent protein, R7 | | Authors: | Hansen, S, Stueber, J, Ernst, P, Koch, A, Bojar, D, Batyuk, A, Plueckthun, A. | | Deposit date: | 2016-11-03 | | Release date: | 2017-11-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Design and applications of a clamp for Green Fluorescent Protein with picomolar affinity.
Sci Rep, 7, 2017
|
|
6NIG
 
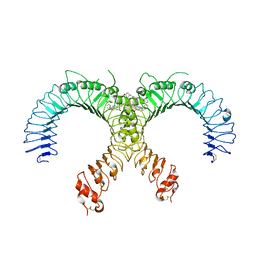 | | Crystal structure of the human TLR2-Diprovocim complex | | Descriptor: | (3S,4S,3'S,4'S)-1,1'-(1,4-phenylenedicarbonyl)bis{N~3~,N~4~-bis[(1S,2R)-2-phenylcyclopropyl]pyrrolidine-3,4-dicarboxami de}, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, H, Beutler, B.A, Tomchick, D.R, Su, L. | | Deposit date: | 2018-12-27 | | Release date: | 2019-04-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Basis of TLR2/TLR1 Activation by the Synthetic Agonist Diprovocim.
J. Med. Chem., 62, 2019
|
|
5MBE
 
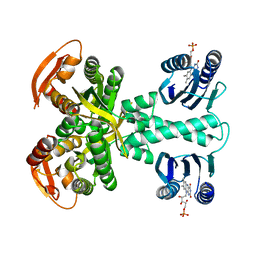 | | Structure of a bacterial light-regulated adenylyl cylcase | | Descriptor: | Beta subunit of photoactivated adenylyl cyclase, FLAVIN MONONUCLEOTIDE | | Authors: | Lindner, R, Hartmann, E, Tarnawski, M, Winkler, A, Frey, D, Reinstein, J, Meinhart, A, Schlichting, I. | | Deposit date: | 2016-11-08 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Photoactivation Mechanism of a Bacterial Light-Regulated Adenylyl Cyclase.
J. Mol. Biol., 429, 2017
|
|
7QOE
 
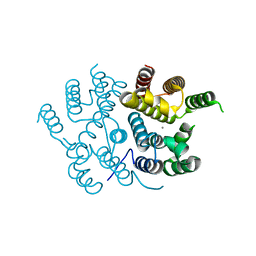 | |
6PPA
 
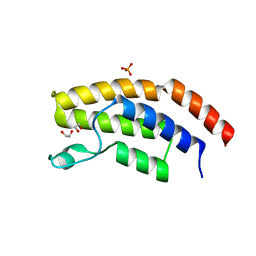 | | Crystal structure of the unliganded bromodomain of human BRD7 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 7, PHOSPHATE ION | | Authors: | Chan, A, Karim, M.R, Zhu, J, Schonbrunn, E. | | Deposit date: | 2019-07-05 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural Basis of Inhibitor Selectivity in the BRD7/9 Subfamily of Bromodomains.
J.Med.Chem., 63, 2020
|
|
8CVI
 
 | |
5MFO
 
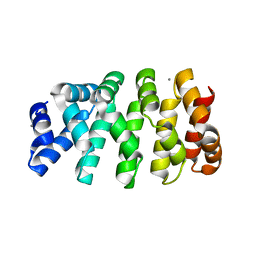 | | Designed armadillo repeat protein YIIIM3AIII | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, YIIIM3AIII | | Authors: | Hansen, S, Ernst, P, Reichen, C, Ewald, C, Mittl, P, Plueckthun, A. | | Deposit date: | 2016-11-18 | | Release date: | 2017-09-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Curvature of designed armadillo repeat proteins allows modular peptide binding.
J. Struct. Biol., 201, 2018
|
|
6VIM
 
 | | P. putida mandelate racemase co-crystallized with phenylboronic acid | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Mandelate racemase, ... | | Authors: | Grandinetti, L, Sharma, A.N, Bearne, S.L, St Maurice, M. | | Deposit date: | 2020-01-13 | | Release date: | 2020-08-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Potent Inhibition of Mandelate Racemase by Boronic Acids: Boron as a Mimic of a Carbon Acid Center.
Biochemistry, 59, 2020
|
|
6YOH
 
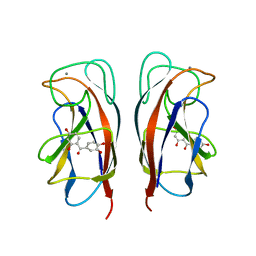 | | LecA from Pseudomonas aeruginosa in complex with a catechol CAS no. 61445-50-9 | | Descriptor: | CALCIUM ION, PA-I galactophilic lectin, [2,4-bis(oxidanyl)phenyl]-[3,4-bis(oxidanyl)phenyl]methanone | | Authors: | Kuhaudomlarp, S, Imberty, A, Titz, A, Varrot, A. | | Deposit date: | 2020-04-14 | | Release date: | 2021-02-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Non-Carbohydrate Glycomimetics as Inhibitors of Calcium(II)-binding Lectins.
Angew.Chem.Int.Ed.Engl., 2020
|
|
6VJP
 
 | | Structure of Staphylococcus aureus peptidoglycan O-acetyltransferase A (OatA) C-terminal catalytic domain | | Descriptor: | Acetyltransferase, SODIUM ION | | Authors: | Jones, C.J, Sychantha, D, Howell, P.L, Clarke, A.J. | | Deposit date: | 2020-01-16 | | Release date: | 2020-05-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.711 Å) | | Cite: | Structural basis for theO-acetyltransferase function of the extracytoplasmic domain of OatA fromStaphylococcus aureus.
J.Biol.Chem., 295, 2020
|
|
6NBG
 
 | | 2.05 Angstrom Resolution Crystal Structure of Hypothetical Protein KP1_5497 from Klebsiella pneumoniae. | | Descriptor: | CHLORIDE ION, Glucosamine-6-phosphate deaminase, PHOSPHATE ION | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-12-07 | | Release date: | 2018-12-19 | | Last modified: | 2023-06-14 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | A Structural Systems Biology Approach to High-Risk CG23 Klebsiella pneumoniae.
Microbiol Resour Announc, 12, 2023
|
|
5MHN
 
 | |
6SUM
 
 | | Amicoumacin kinase hAmiN in complex with AMP-PNP, MG2+ and Ami | | Descriptor: | ACETATE ION, AMICOUMACIN KINASE, Amicoumacin A, ... | | Authors: | Bourenkov, G.P, Mokrushina, Y.A, Terekhov, S.S, Smirnov, I.V, Gabibov, A.G, Altman, S. | | Deposit date: | 2019-09-16 | | Release date: | 2020-07-22 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | A kinase bioscavenger provides antibiotic resistance by extremely tight substrate binding.
Sci Adv, 6, 2020
|
|
7A0L
 
 | | Joint neutron/X-ray room temperature structure of perdeuterated Aspergillus flavus urate oxidase in complex with the 8-azaxanthine inhibitor and catalytic water bound in the peroxo hole | | Descriptor: | 8-AZAXANTHINE, SODIUM ION, Uricase | | Authors: | McGregor, L, Bui, S, Blakeley, M.P, Steiner, R.A. | | Deposit date: | 2020-08-09 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-01 | | Method: | NEUTRON DIFFRACTION (1.33 Å), X-RAY DIFFRACTION | | Cite: | Joint neutron/X-ray crystal structure of a mechanistically relevant complex of perdeuterated urate oxidase and simulations provide insight into the hydration step of catalysis.
Iucrj, 8, 2021
|
|
6YC6
 
 | | Structure of C. glutamicum GlnK | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHATE ION, PII protein | | Authors: | Grau, F.C, Muller, Y.A. | | Deposit date: | 2020-03-18 | | Release date: | 2021-03-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of adenylylated and unadenylylated P II protein GlnK from Corynebacterium glutamicum.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
