8QOG
 
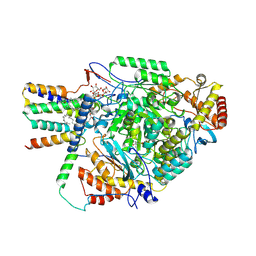 | | Cryo-EM structure of the yeast SPT-Orm2-Monomer complex | | Descriptor: | 2-{[(4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranosyl)oxy]methyl}-4-{[(3beta,9beta,14beta,17beta,25R)-spirost-5-en-3-yl]oxy}butyl 4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranoside, ORM2 isoform 1, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Schaefer, J, Koerner, C, Moeller, A, Froehlich, F. | | Deposit date: | 2023-09-28 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The structure of the Orm2-containing serine palmitoyltransferase complex reveals distinct inhibitory potentials of yeast Orm proteins.
Cell Rep, 43, 2024
|
|
6F9F
 
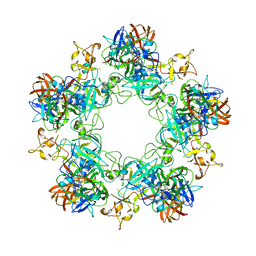 | |
4YG8
 
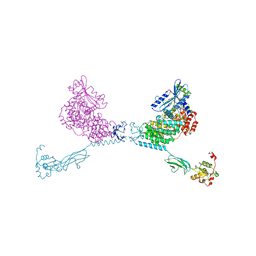 | | CRYSTAL STRUCTURE OF THE CHS5-CHS6 EXOMER CARGO ADAPTOR COMPLEX | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Chitin biosynthesis protein CHS5, Chitin biosynthesis protein CHS6, ... | | Authors: | Paczkowski, J.E, Richardson, B.C, Strassner, A.M, Fromme, J.C. | | Deposit date: | 2015-02-26 | | Release date: | 2015-03-11 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The exomer cargo adaptor structure reveals a novel GTPase-binding domain.
Embo J., 31, 2012
|
|
5C2G
 
 | | GWS1B RubisCO: Form II RubisCO derived from uncultivated Gallionellacea species (CABP-bound). | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, Form II RubisCO, MAGNESIUM ION | | Authors: | Arbing, M.A, Varaljay, V.A, Satagopan, S, Tabita, F.R. | | Deposit date: | 2015-06-15 | | Release date: | 2015-12-16 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (2.597 Å) | | Cite: | Functional metagenomic selection of ribulose 1, 5-bisphosphate carboxylase/oxygenase from uncultivated bacteria.
Environ.Microbiol., 18, 2016
|
|
4PYL
 
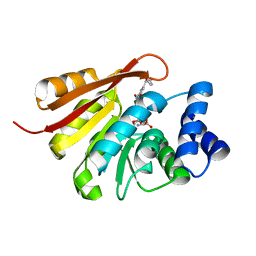 | | Humanized rat COMT in complex with sinefungin, Mg2+, and tolcapone | | Descriptor: | Catechol O-methyltransferase, MAGNESIUM ION, SINEFUNGIN, ... | | Authors: | Ehler, A, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2014-03-27 | | Release date: | 2014-06-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mapping the conformational space accessible to catechol-O-methyltransferase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4Y9V
 
 | | Gp54 tailspike of Acinetobacter baumannii bacteriophage AP22 in complex with A. baumannii capsular saccharide | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2,4-dideoxy-alpha-L-erythro-hex-4-enopyranuronic acid-(1-3)-2-acetamido-2-deoxy-alpha-D-fucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-mannopyranuronic acid, CHLORIDE ION, ... | | Authors: | Buth, S.A, Shneider, M.M, Leiman, P.G. | | Deposit date: | 2015-02-17 | | Release date: | 2017-02-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Structure of Acinetobacter baumannii bacteriophage AP22 polysaccharide degrading lyase in complex with A. baumannii capsular saccharide at 0.9 A resolution
TO BE PUBLISHED
|
|
6FBR
 
 | | Crystal Structure of the Human Retinoid X Receptor DNA-Binding Domain Bound to the Human MEp DR1 Response Element, pH 4.2 | | Descriptor: | DI(HYDROXYETHYL)ETHER, DNA (5'-D(*CP*TP*GP*GP*GP*TP*CP*AP*AP*AP*GP*TP*TP*CP*AP*TP*C)-3'), DNA (5'-D(*GP*AP*TP*GP*AP*AP*CP*TP*TP*TP*GP*AP*CP*CP*CP*AP*G)-3'), ... | | Authors: | McEwen, A.G, Poussin-Courmontagne, P, Osz, J, Rochel, N. | | Deposit date: | 2017-12-19 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Modulation of RXR-DNA complex assembly by DNA context.
Mol. Cell. Endocrinol., 481, 2019
|
|
5U7A
 
 | | Crystal structure of a complex formed between MerB and Dimethyltin | | Descriptor: | Alkylmercury lyase, BROMIDE ION, Dimethyltin dibromide, ... | | Authors: | Wahba, H.M, Stevenson, M, Mansour, A, Sygusch, J, Wilcox, D.E, Omichinski, J.G. | | Deposit date: | 2016-12-12 | | Release date: | 2017-01-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.532 Å) | | Cite: | Structural and Biochemical Characterization of Organotin and Organolead Compounds Binding to the Organomercurial Lyase MerB Provide New Insights into Its Mechanism of Carbon-Metal Bond Cleavage.
J. Am. Chem. Soc., 139, 2017
|
|
5C6J
 
 | | Crystal Structure of Gadolinium derivative of HEWL solved using Free-Electron Laser radiation | | Descriptor: | 10-((2R)-2-HYDROXYPROPYL)-1,4,7,10-TETRAAZACYCLODODECANE 1,4,7-TRIACETIC ACID, CHLORIDE ION, GADOLINIUM ATOM, ... | | Authors: | Galli, L, Barends, T.R.M, Son, S.-K, White, T.A, Barty, A, Botha, S, Boutet, S, Caleman, C, Doak, R.B, Nanao, M.H, Nass, K, Shoeman, R.L, Timneanu, N, Santra, R, Schlichting, I, Chapman, H.N. | | Deposit date: | 2015-06-23 | | Release date: | 2015-07-08 | | Last modified: | 2018-11-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Towards phasing using high X-ray intensity.
Iucrj, 2, 2015
|
|
6FJT
 
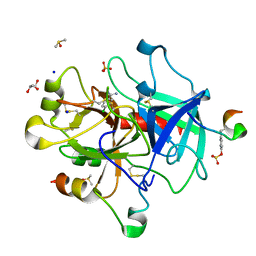 | | 4-chloro-benzamidine in complex with thrombin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-chloranylbenzenecarboximidamide, DIMETHYL SULFOXIDE, ... | | Authors: | Abazi, N, Heine, A, Klebe, G. | | Deposit date: | 2018-01-23 | | Release date: | 2019-02-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | 4-chloro-benzamidine in complex with thrombin
To be published
|
|
6CQX
 
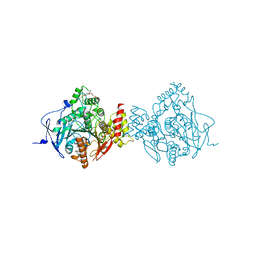 | | Crystal Structure of Recombinant Human Acetylcholinesterase Inhibited by VX(+) | | Descriptor: | 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bester, S.M, Guelta, M.A, Pegan, S.D, Height, J.J. | | Deposit date: | 2018-03-16 | | Release date: | 2018-12-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Insights of Stereospecific Inhibition of Human Acetylcholinesterase by VX and Subsequent Reactivation by HI-6.
Chem. Res. Toxicol., 31, 2018
|
|
4YLH
 
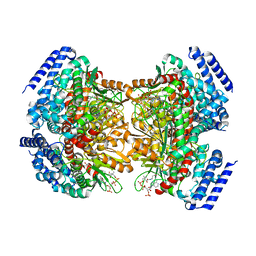 | | Crystal structure of DpgC with bound substrate analog and Xe on oxygen diffusion pathway | | Descriptor: | DpgC, XENON, [(2R,3S,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-4-HYDROXY-3-(PHOSPHONOOXY)TETRAHYDROFURAN-2-YL]METHYL (3R)-4-({3-[(2-{[(3,5-DIHYDROXYPHENYL)ACETYL]AMINO}ETHYL)AMINO]-3-OXOPROPYL}AMINO)-3-HYDROXY-2,2-DIMETHYL-4-OXOBUTYL DIHYDROGEN DIPHOSPHATE | | Authors: | Li, K, Di Russo, N.V, Condurso, H.L, Roitberg, A.E, Bruner, S.D. | | Deposit date: | 2015-03-05 | | Release date: | 2015-08-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Oxygen diffusion pathways in a cofactor-independent dioxygenase.
Chem Sci, 6, 2015
|
|
5TUD
 
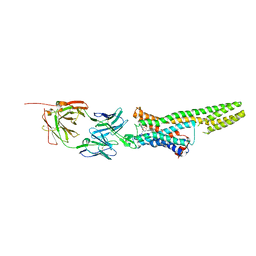 | | Structural Insights into the Extracellular Recognition of the Human Serotonin 2B Receptor by an Antibody | | Descriptor: | 5-hydroxytryptamine receptor 2B,Soluble cytochrome b562 chimera, Anti-5-HT2B Fab heavy chain, Anti-5-HT2B Fab light chain, ... | | Authors: | Ishchenko, A, Wacker, D, Kapoor, M, Zhang, A, Han, G.W, Basu, S, Boutet, S, James, D, Wang, D, Weierstall, U, Liu, W, Katritch, V, Stevens, R.C, Cherezov, V. | | Deposit date: | 2016-11-05 | | Release date: | 2017-07-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural insights into the extracellular recognition of the human serotonin 2B receptor by an antibody.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6J2M
 
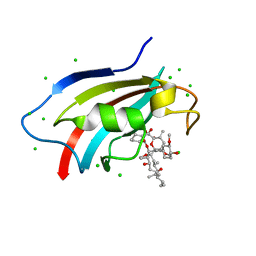 | | Crystal structure of AtFKBP53 C-terminal domain | | Descriptor: | 8-DEETHYL-8-[BUT-3-ENYL]-ASCOMYCIN, CHLORIDE ION, Peptidyl-prolyl cis-trans isomerase FKBP53 | | Authors: | Singh, A.K, Vasudevan, D. | | Deposit date: | 2019-01-01 | | Release date: | 2019-12-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | AtFKBP53: a chimeric histone chaperone with functional nucleoplasmin and PPIase domains.
Nucleic Acids Res., 48, 2020
|
|
5TW4
 
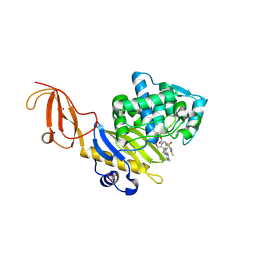 | |
7AHW
 
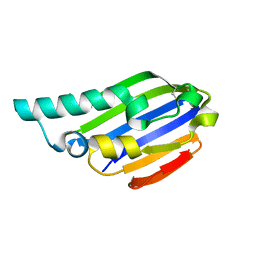 | |
5CGB
 
 | | Crystal structure of FimH in complex with heptyl alpha-D-septanoside | | Descriptor: | (6R)-1,6-anhydro-2-O-heptyl-6-(hydroxymethyl)-D-galactitol, Protein FimH, SULFATE ION | | Authors: | Jakob, R.P, Preston, R.P, Zihlmann, P, Fiege, B, Sager, C.P, Vannam, R, Rabbani, S, Zalewski, A, Maier, T, Ernst, B, Peczuh, M. | | Deposit date: | 2015-07-09 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The price of flexibility - a case study on septanoses as pyranose mimetics.
Chem Sci, 9, 2018
|
|
5CCL
 
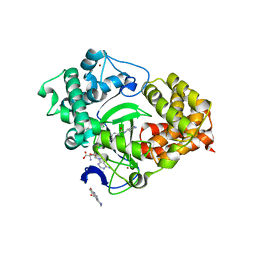 | | Crystal structure of SMYD3 with SAM and oxindole compound | | Descriptor: | 1,2-ETHANEDIOL, 2-oxidanylidene-N-piperidin-4-yl-1,3-dihydroindole-5-carboxamide, Histone-lysine N-methyltransferase SMYD3, ... | | Authors: | Boriack-Sjodin, P.A. | | Deposit date: | 2015-07-02 | | Release date: | 2015-09-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Novel Oxindole Sulfonamides and Sulfamides: EPZ031686, the First Orally Bioavailable Small Molecule SMYD3 Inhibitor.
Acs Med.Chem.Lett., 7, 2016
|
|
7KP5
 
 | |
6SBL
 
 | | Human Carbonic Anhydrase II in complex with 4-hexylbenzenesulfonamide | | Descriptor: | (4-CARBOXYPHENYL)(CHLORO)MERCURY, 4-hexylbenzenesulfonamide, CITRATE ANION, ... | | Authors: | Gloeckner, S, Ngo, K, Heine, A, Klebe, G. | | Deposit date: | 2019-07-21 | | Release date: | 2020-08-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (0.94 Å) | | Cite: | Human Carbonic Anhydrase II in complex with 4-hexylbenzenesulfonamide
To Be Published
|
|
6SBX
 
 | | CdbA Form Two | | Descriptor: | CdbA | | Authors: | Lovering, A.L, Cadby, I.T. | | Deposit date: | 2019-07-22 | | Release date: | 2020-04-22 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | CdbA is a DNA-binding protein and c-di-GMP receptor important for nucleoid organization and segregation in Myxococcus xanthus.
Nat Commun, 11, 2020
|
|
6CLC
 
 | | 1.01 A MicroED structure of GSNQNNF at 0.27 e- / A^2 | | Descriptor: | ACETATE ION, GSNQNNF, ZINC ION | | Authors: | Hattne, J, Shi, D, Glynn, C, Zee, C.-T, Gallagher-Jones, M, Martynowycz, M.W, Rodriguez, J.A, Gonen, T. | | Deposit date: | 2018-03-02 | | Release date: | 2018-05-16 | | Last modified: | 2024-03-13 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.01 Å) | | Cite: | Analysis of Global and Site-Specific Radiation Damage in Cryo-EM.
Structure, 26, 2018
|
|
4YQL
 
 | | Crystal structure of TrmD, a M1G37 tRNA Methyltransferase with SAM-competitive compounds | | Descriptor: | 4-amino-N-[(1S,2R)-2-(4-sulfamoylphenyl)cyclopropyl]-1,2,5-oxadiazole-3-carboxamide, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Elkins, P.A, Bonnette, W.G, Madauss, K.P. | | Deposit date: | 2015-03-13 | | Release date: | 2016-03-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Crystal structure of TrmD, a M1G37 tRNA Methyltransferase with SAM-competitive compounds
To Be Published
|
|
6SCG
 
 | | Structure of AdhE form 1 | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Aldehyde-alcohol dehydrogenase, ... | | Authors: | Lovering, A.L, Bragginton, E. | | Deposit date: | 2019-07-24 | | Release date: | 2020-08-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | High-resolution structure of the alcohol dehydrogenase domain of the bifunctional bacterial enzyme AdhE.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
6CLH
 
 | | 1.37 A MicroED structure of GSNQNNF at 2.9 e- / A^2 | | Descriptor: | ACETATE ION, GSNQNNF, ZINC ION | | Authors: | Hattne, J, Shi, D, Glynn, C, Zee, C.-T, Gallagher-Jones, M, Martynowycz, M.W, Rodriguez, J.A, Gonen, T. | | Deposit date: | 2018-03-02 | | Release date: | 2018-05-16 | | Last modified: | 2024-03-13 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.37 Å) | | Cite: | Analysis of Global and Site-Specific Radiation Damage in Cryo-EM.
Structure, 26, 2018
|
|
