6BVC
 
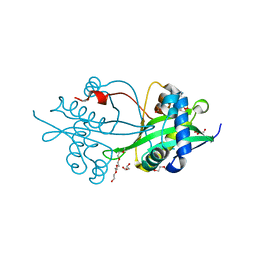 | | Crystal structure of AAC(3)-Ia in complex with coenzyme A | | Descriptor: | 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, Aminoglycoside-(3)-N-acetyltransferase, CHLORIDE ION, ... | | Authors: | Stogios, P.J, Evdokimova, E, Wawrzak, Z, Savchenko, A, Joachimiak, A, Satchell, K, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-12-12 | | Release date: | 2017-12-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.808 Å) | | Cite: | To be published
To Be Published
|
|
6E4Q
 
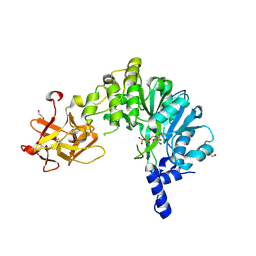 | | Crystal Structure of the Drosophila Melanogaster Polypeptide N-Acetylgalactosaminyl Transferase PGANT9A in Complex with UDP and Mn2+ | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Samara, N.L, Tabak, L.A, Ten Hagen, K.G. | | Deposit date: | 2018-07-18 | | Release date: | 2018-09-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.799 Å) | | Cite: | A molecular switch orchestrates enzyme specificity and secretory granule morphology.
Nat Commun, 9, 2018
|
|
4ZGX
 
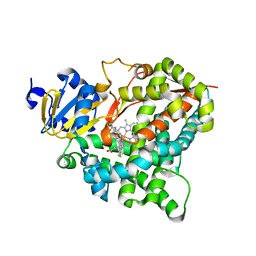 | | Structure of aldosterone synthase (CYP11B2) in complex with (+)-(R)-N-(4-(4-chloro-3-fluorophenyl)-5,6,7,8-tetrahydroisoquinolin-8-yl)propionamide | | Descriptor: | Cytochrome P450 11B2, mitochondrial, N-[(8R)-4-(4-chloro-3-fluorophenyl)-5,6,7,8-tetrahydroisoquinolin-8-yl]propanamide, ... | | Authors: | Kuglstatter, A, Joseph, C. | | Deposit date: | 2015-04-24 | | Release date: | 2015-10-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Discovery of 4-Aryl-5,6,7,8-tetrahydroisoquinolines as Potent, Selective, and Orally Active Aldosterone Synthase (CYP11B2) Inhibitors: In Vivo Evaluation in Rodents and Cynomolgus Monkeys.
J.Med.Chem., 58, 2015
|
|
5AOT
 
 | | Very high resolution structure of a novel carbohydrate binding module from Ruminococcus flavefaciens FD-1 endoglucanase Cel5A | | Descriptor: | CACODYLATE ION, Carbohydrate binding module, GLYCEROL | | Authors: | Pires, A.J, Ribeiro, T, Thompson, A, Venditto, I, Fernandes, V.O, Bule, P, Santos, H, Alves, V.D, Pires, V, Ferreira, L.M.A, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2015-09-11 | | Release date: | 2016-06-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Complexity of the Ruminococcus flavefaciens cellulosome reflects an expansion in glycan recognition.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
6E55
 
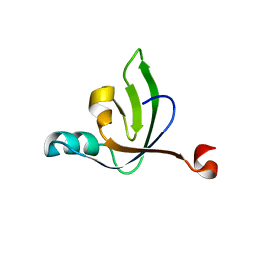 | |
1AEF
 
 | | SPECIFICITY OF LIGAND BINDING TO A BURIED POLAR CAVITY AT THE ACTIVE SITE OF CYTOCHROME C PEROXIDASE (3-AMINOPYRIDINE) | | Descriptor: | 3-AMINOPYRIDINE, CYTOCHROME C PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Musah, R.A, Jensen, G.M, Fitzgerald, M.M, Mcree, D.E, Goodin, D.B. | | Deposit date: | 1997-02-24 | | Release date: | 1997-09-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Artificial protein cavities as specific ligand-binding templates: characterization of an engineered heterocyclic cation-binding site that preserves the evolved specificity of the parent protein.
J.Mol.Biol., 315, 2002
|
|
4U01
 
 | | HCV NS3/4A serine protease in complex with 6570 | | Descriptor: | (2S,3aS,10Z,11aS,12aR)-2-({8-fluoro-7-methoxy-2-[4-(propan-2-yl)-1,3-thiazol-2-yl]quinolin-4-yl}oxy)-5-methyl-N-[(1-methylcyclopropyl)sulfonyl]-4,14-dioxo-1,2,3,3a,4,5,6,7,8,9,11a,12,13,14-tetradecahydro-12aH-cyclopropa[m]pyrrolo[1,2-c][1,3,6]triazacyclotetradecine-12a-carboxamide, CHLORIDE ION, NS4A protein, ... | | Authors: | Parsy, C.C, Alexandre, F.-R, Brandt, G, Caillet, C, Chaves, D, Derock, M, Gloux, D, Griffon, Y, Lallos, L.B, Leroy, F, Liuzzi, M, Loi, A.-G, Mayes, B, Moulat, L, Moussa, A, Chiara, M, Roques, V, Rosinovsky, E, Seifer, M, Stewart, A, Wang, J, Standring, D, Surleraux, D. | | Deposit date: | 2014-07-11 | | Release date: | 2015-07-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery and structural diversity of the hepatitis C virus NS3/4A serine protease inhibitor series leading to clinical candidate IDX320.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
6E21
 
 | | Joint X-ray/neutron structure of PKAc with products Sr2-ADP and phosphorylated peptide SP20 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, STRONTIUM ION, cAMP-dependent protein kinase catalytic subunit alpha, ... | | Authors: | Kovalevsky, A, Gerlits, O.O, Taylor, S. | | Deposit date: | 2018-07-10 | | Release date: | 2019-04-03 | | Last modified: | 2023-10-25 | | Method: | NEUTRON DIFFRACTION (2 Å), X-RAY DIFFRACTION | | Cite: | Zooming in on protons: Neutron structure of protein kinase A trapped in a product complex.
Sci Adv, 5, 2019
|
|
6H8L
 
 | | Structure of peptidoglycan deacetylase PdaC from Bacillus subtilis | | Descriptor: | L(+)-TARTARIC ACID, Peptidoglycan-N-acetylmuramic acid deacetylase PdaC, ZINC ION | | Authors: | Sainz-Polo, M.A, Grifoll-Romero, L, Albesa-Jove, D, Planas, A, Guerin, M.E. | | Deposit date: | 2018-08-02 | | Release date: | 2019-11-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structure-function relationships underlying the dualN-acetylmuramic andN-acetylglucosamine specificities of the bacterial peptidoglycan deacetylase PdaC.
J.Biol.Chem., 294, 2019
|
|
5HW7
 
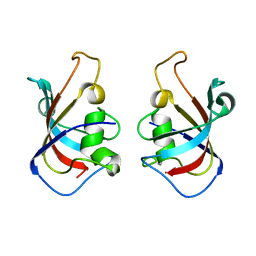 | |
1AGY
 
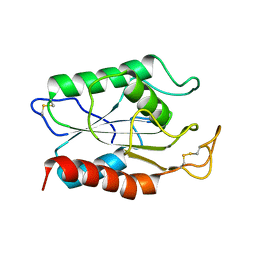 | |
5V3J
 
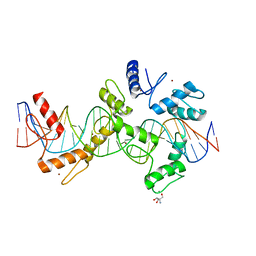 | | mouseZFP568-ZnF1-10 in complex with DNA | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DNA (26-MER), MAGNESIUM ION, ... | | Authors: | Patel, A, Cheng, X. | | Deposit date: | 2017-03-07 | | Release date: | 2018-03-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.064 Å) | | Cite: | DNA Conformation Induces Adaptable Binding by Tandem Zinc Finger Proteins.
Cell, 173, 2018
|
|
1A4A
 
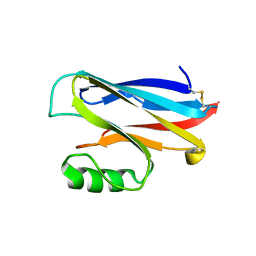 | |
7ZJY
 
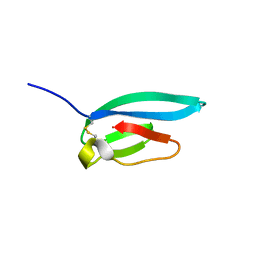 | | The NMR structure of the MAX67 effector from Magnaporthe Oryzae | | Descriptor: | MAX effector protein | | Authors: | Lahfa, M, Padilla, A, de Guillen, K, Pissarra, J, Raji, M, Cesari, S, Kroj, T, Gladieux, P, Roumestand, C, Barthe, P. | | Deposit date: | 2022-04-12 | | Release date: | 2023-04-26 | | Last modified: | 2024-09-04 | | Method: | SOLUTION NMR | | Cite: | The structural landscape and diversity of Pyricularia oryzae MAX effectors revisited.
Plos Pathog., 20, 2024
|
|
5JKI
 
 | | Crystal structure of the first transmembrane PAP2 type phosphatidylglycerolphosphate phosphatase from Bacillus subtilis | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Putative lipid phosphate phosphatase YodM, TUNGSTATE(VI)ION, ... | | Authors: | El Ghachi, M, Howe, N, Lampion, A, Delbrassine, F, Vogeley, L, Caffrey, M, Sauvage, E, Auger, R, Guiseppe, A, Roure, S, Perlier, S, Mengin-lecreulx, D, Foglino, M, Touze, T. | | Deposit date: | 2016-04-26 | | Release date: | 2017-02-22 | | Last modified: | 2017-05-24 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure and biochemical characterization of the transmembrane PAP2 type phosphatidylglycerol phosphate phosphatase from Bacillus subtilis.
Cell. Mol. Life Sci., 74, 2017
|
|
7ZKA
 
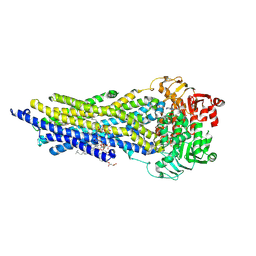 | | ABCB1 V978C mutant (mABCB1) in the outward facing state bound to AAC | | Descriptor: | (4~{S},11~{S},18~{S})-4,11-dimethyl-18-(sulfanylmethyl)-6,13,20-trithia-3,10,17,22,23,24-hexazatetracyclo[17.2.1.1^{5,8}.1^{12,15}]tetracosa-1(21),5(24),7,12(23),14,19(22)-hexaene-2,9,16-trione, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent translocase ABCB1, ... | | Authors: | Parey, K, Januliene, D, Gewering, T, Urbatsch, I, Zhang, Q, Moeller, A. | | Deposit date: | 2022-04-12 | | Release date: | 2023-04-26 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Tracing the substrate translocation mechanism in P-glycoprotein.
Elife, 12, 2024
|
|
5JLR
 
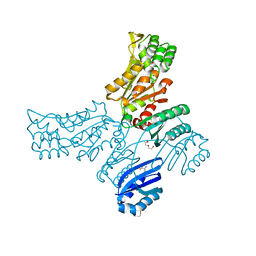 | | Crystal structure of Mycobacterium avium SerB2 with serine present at slightly different position near ACT domain | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-METHOXYETHOXY)ETHANOL, MAGNESIUM ION, ... | | Authors: | Shree, S, Agrawal, A, Dubey, S, Ramachandran, R. | | Deposit date: | 2016-04-27 | | Release date: | 2017-05-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.261 Å) | | Cite: | Crystal structure of Mycobacterium avium SerB2 with serine present at slightly different position near ACT domain
To be published
|
|
6HAU
 
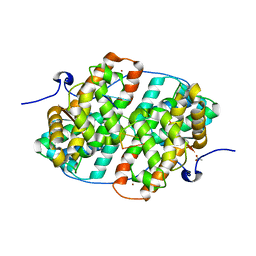 | | KSHV PAN RNA Mta-response element fragment complexed with the globular domain of herpesvirus saimiri ORF57 | | Descriptor: | ACETATE ION, MRE fragment of PAN RNA, ZINC ION, ... | | Authors: | Tunnicliffe, R.B, Levy, C, Ruiz Nivia, H.D, Sandri-Goldin, R.M, Golovanov, A.P. | | Deposit date: | 2018-08-08 | | Release date: | 2018-11-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural identification of conserved RNA binding sites in herpesvirus ORF57 homologs: implications for PAN RNA recognition.
Nucleic Acids Res., 47, 2019
|
|
7ZK0
 
 | | The NMR structure of the MAX60 effector from Magnaporthe Oryzae | | Descriptor: | MAX effector protein | | Authors: | Lahfa, M, Padilla, A, de Guillen, K, Pissarra, J, Raji, M, Cesari, S, Kroj, T, Gladieux, P, Roumestand, C, Barthe, P. | | Deposit date: | 2022-04-12 | | Release date: | 2023-04-26 | | Last modified: | 2024-09-04 | | Method: | SOLUTION NMR | | Cite: | The structural landscape and diversity of Pyricularia oryzae MAX effectors revisited.
Plos Pathog., 20, 2024
|
|
7ZK6
 
 | | ABCB1 L335C mutant (mABCB1) in the outward facing state bound to 2 molecules of AAC | | Descriptor: | (4S,11S,18S)-4-[[(2,4-dinitrophenyl)disulfanyl]methyl]-11,18-dimethyl-6,13,20-trithia-3,10,17,22,23,24-hexazatetracyclo[17.2.1.1^{5,8}.1^{12,15}]tetracosa-1(21),5(24),7,12(23),14,19(22)-hexaene-2,9,16-trione, (4~{S},11~{S},18~{S})-4,11-dimethyl-18-(sulfanylmethyl)-6,13,20-trithia-3,10,17,22,23,24-hexazatetracyclo[17.2.1.1^{5,8}.1^{12,15}]tetracosa-1(21),5(24),7,12(23),14,19(22)-hexaene-2,9,16-trione, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Parey, K, Januliene, D, Gewering, T, Moeller, A. | | Deposit date: | 2022-04-12 | | Release date: | 2023-04-26 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Tracing the substrate translocation mechanism in P-glycoprotein.
Elife, 12, 2024
|
|
6W9T
 
 | |
5JM1
 
 | | Structure of tetrameric jacalin complexed with a trisaccharide, Gal alpha-(1,3) Gal beta-(1,4) Gal | | Descriptor: | 1,2-ETHANEDIOL, Agglutinin alpha chain, Agglutinin beta-3 chain, ... | | Authors: | Abhinav, K.V, Sharma, K, Surolia, A, Vijayan, M. | | Deposit date: | 2016-04-28 | | Release date: | 2016-12-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Effect of linkage on the location of reducing and nonreducing sugars bound to jacalin.
IUBMB Life, 68, 2016
|
|
5HZC
 
 | | Crystal structure of the complex PPARgamma/AL26-29 | | Descriptor: | 2-methyl-2-[4-(naphthalen-1-yl)phenoxy]propanoic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Pochetti, G, Montanari, R, Capelli, D, Loiodice, F, Laghezza, A, Lavecchia, A. | | Deposit date: | 2016-02-02 | | Release date: | 2016-11-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for PPAR partial or full activation revealed by a novel ligand binding mode.
Sci Rep, 6, 2016
|
|
6YUE
 
 | | Fragment of nitrate/nitrite sensor histidine kinase NarQ (R50S variant) | | Descriptor: | Nitrate/nitrite sensor protein NarQ | | Authors: | Gushchin, I, Melnikov, I, Polovinkin, V, Yuzhakova, A, Gordeliy, V. | | Deposit date: | 2020-04-27 | | Release date: | 2020-05-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Sensor Histidine Kinase NarQ Activates via Helical Rotation, Diagonal Scissoring, and Eventually Piston-Like Shifts.
Int J Mol Sci, 21, 2020
|
|
7ZK5
 
 | | ABCB1 L335C mutant (mABCB1) in the outward facing state bound to AAC | | Descriptor: | (4~{S},11~{S},18~{S})-4,11-dimethyl-18-(sulfanylmethyl)-6,13,20-trithia-3,10,17,22,23,24-hexazatetracyclo[17.2.1.1^{5,8}.1^{12,15}]tetracosa-1(21),5(24),7,12(23),14,19(22)-hexaene-2,9,16-trione, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent translocase ABCB1, ... | | Authors: | Parey, K, Januliene, D, Gewering, T, Moeller, A. | | Deposit date: | 2022-04-12 | | Release date: | 2023-04-26 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Tracing the substrate translocation mechanism in P-glycoprotein.
Elife, 12, 2024
|
|
