1XEE
 
 | | Solution structure of the Chemotaxis Inhibitory Protein of Staphylococcus aureus | | Descriptor: | chemotaxis-inhibiting protein CHIPS | | Authors: | Haas, P.J, de Haas, C.J, Poppelier, M.J, van Kessel, K.P, van Strijp, J.A, Dijkstra, K, Scheek, R.M, Fan, H, Kruijtzer, J.A, Liskamp, R.M, Kemmink, J. | | Deposit date: | 2004-09-10 | | Release date: | 2005-09-27 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The structure of the C5a receptor-blocking domain of chemotaxis inhibitory protein of Staphylococcus aureus is related to a group of immune evasive molecules
J.Mol.Biol., 353, 2005
|
|
4NZL
 
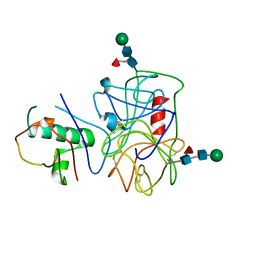 | | Extracellular proteins of Staphylococcus aureus inhibit the neutrophil serine proteases | | Descriptor: | Neutrophil elastase, Uncharacterized protein, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Stapels, D.A.C, von Koeckritz-Blickwede, M, Ramyar, K.X, Bischoff, M, Milder, F, Ruyken, M, Scheepmaker, L, McWhorter, W.J, Herrmann, M, van Kessel, K.P.M, Geisbrecht, B.V, Rooijakkers, S.H.M. | | Deposit date: | 2013-12-12 | | Release date: | 2014-08-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Staphylococcus aureus secretes a unique class of neutrophil serine protease inhibitors.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
5D3D
 
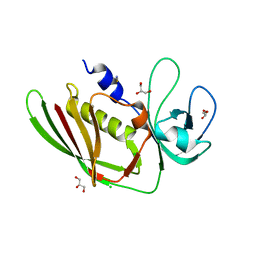 | |
5D3I
 
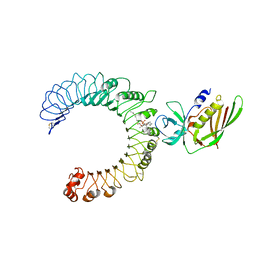 | | Crystal structure of the SSL3-TLR2 complex | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Feitsma, L.J, Huizinga, E.G. | | Deposit date: | 2015-08-06 | | Release date: | 2015-08-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for inhibition of TLR2 by staphylococcal superantigen-like protein 3 (SSL3).
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5UZU
 
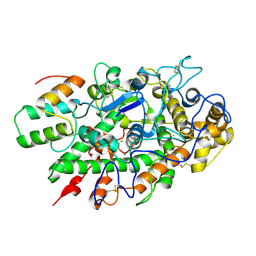 | | Immune evasion by a Staphylococcal Peroxidase Inhibitor that blocks myeloperoxidase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | de Jong, N, Geisbrecht, B.V, van Strijp, J, Haas, P, Nijland, R, Ramyar, K, Fevre, C, Guerra, F, Voyich-Kane, J, van Kessel, C, Garcia, B. | | Deposit date: | 2017-02-27 | | Release date: | 2017-07-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.403 Å) | | Cite: | Immune evasion by a staphylococcal inhibitor of myeloperoxidase.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6AZP
 
 | |
6BMT
 
 | |
