1QB2
 
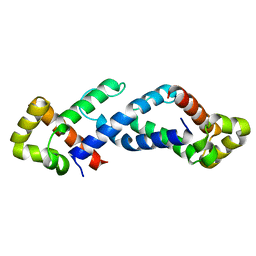 | | CRYSTAL STRUCTURE OF THE CONSERVED SUBDOMAIN OF HUMAN PROTEIN SRP54M AT 2.1A RESOLUTION: EVIDENCE FOR THE MECHANISM OF SIGNAL PEPTIDE BINDING | | Descriptor: | HUMAN SIGNAL RECOGNITION PARTICLE 54 KD PROTEIN | | Authors: | Clemons Jr, W.M, Gowda, K, Black, S.D, Zwieb, C, Ramakrishnan, V. | | Deposit date: | 1999-04-29 | | Release date: | 1999-10-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the conserved subdomain of human protein SRP54M at 2.1 A resolution: evidence for the mechanism of signal peptide binding.
J.Mol.Biol., 292, 1999
|
|
2JQE
 
 | |
1KVV
 
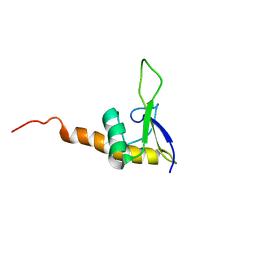 | | Solution Structure Of Protein SRP19 Of The Archaeoglobus fulgidus Signal Recognition Particle, Minimized Average Structure | | Descriptor: | SRP19 | | Authors: | Pakhomova, O.N, Deep, S, Huang, Q, Zwieb, C, Hinck, A.P. | | Deposit date: | 2002-01-27 | | Release date: | 2002-03-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of protein SRP19 of Archaeoglobus fulgidus signal recognition particle.
J.Mol.Biol., 317, 2002
|
|
1KVN
 
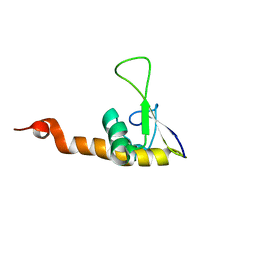 | | Solution Structure Of Protein SRP19 Of The Arhaeoglobus fulgidus Signal Recognition Particle, 10 Structures | | Descriptor: | SRP19 | | Authors: | Pakhomova, O.N, Deep, S, Huang, Q, Zwieb, C, Hinck, A.P. | | Deposit date: | 2002-01-27 | | Release date: | 2002-03-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of protein SRP19 of Archaeoglobus fulgidus signal recognition particle.
J.Mol.Biol., 317, 2002
|
|
3IZ4
 
 | |
6MZP
 
 | |
6MZN
 
 | |
1ZC8
 
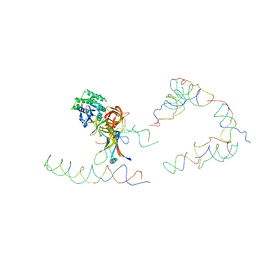 | | Coordinates of tmRNA, SmpB, EF-Tu and h44 fitted into Cryo-EM map of the 70S ribosome and tmRNA complex | | Descriptor: | Elongation factor Tu, H2 16S rRNA, H2b d mRNA, ... | | Authors: | Valle, M, Gillet, R, Kaur, S, Henne, A, Ramakrishnan, V, Frank, J. | | Deposit date: | 2005-04-11 | | Release date: | 2005-04-19 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (13 Å) | | Cite: | Visualizing tmRNA Entry into a Stalled Ribosome
Science, 300, 2003
|
|
