3JB2
 
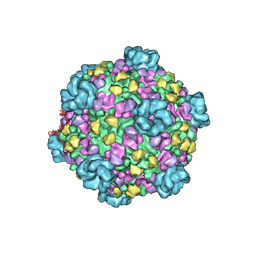 | | Atomic model of cytoplasmic polyhedrosis virus with SAM and GTP | | Descriptor: | Capsid protein VP1, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Yu, X.K, Jiang, J.S, Sun, J.C, Zhou, Z.H. | | Deposit date: | 2015-07-06 | | Release date: | 2015-08-12 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | A putative ATPase mediates RNA transcription and capping in a dsRNA virus.
Elife, 4, 2015
|
|
3J2V
 
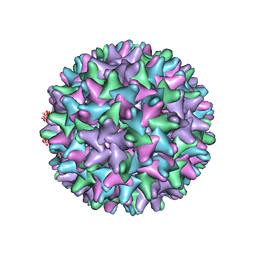 | | CryoEM structure of HBV core | | Descriptor: | PreC/core protein | | Authors: | Yu, X, Jin, L, Jih, J, Shih, C, Zhou, Z.H. | | Deposit date: | 2013-01-11 | | Release date: | 2013-10-02 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | 3.5 angstrom cryoEM Structure of Hepatitis B Virus Core Assembled from Full-Length Core Protein.
Plos One, 8, 2013
|
|
3J9R
 
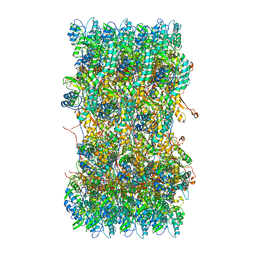 | | Atomic structures of a bactericidal contractile nanotube in its pre- and post-contraction states | | Descriptor: | sheath | | Authors: | Ge, P, Scholl, D, Leiman, P.G, Yu, X, Miller, J.F, Zhou, Z.H. | | Deposit date: | 2015-02-17 | | Release date: | 2015-04-01 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Atomic structures of a bactericidal contractile nanotube in its pre- and postcontraction states.
Nat.Struct.Mol.Biol., 22, 2015
|
|
3JAY
 
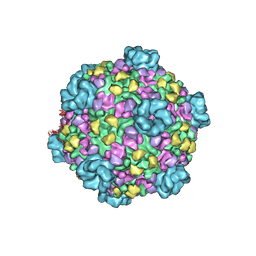 | | Atomic model of transcribing cytoplasmic polyhedrosis virus | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Capsid protein VP1, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Yu, X.K, Jiang, J.S, Sun, J.C, Zhou, Z.H. | | Deposit date: | 2015-07-06 | | Release date: | 2015-08-12 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A putative ATPase mediates RNA transcription and capping in a dsRNA virus.
Elife, 4, 2015
|
|
3JB1
 
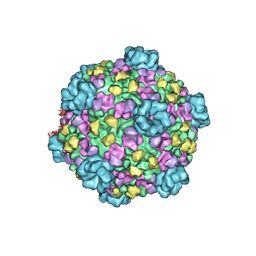 | | Atomic model of cytoplasmic polyhedrosis virus with SAM | | Descriptor: | Capsid protein VP1, S-ADENOSYLMETHIONINE, Structural protein VP3, ... | | Authors: | Yu, X.K, Jiang, J.S, Sun, J.C, Zhou, Z.H. | | Deposit date: | 2015-07-06 | | Release date: | 2015-08-12 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | A putative ATPase mediates RNA transcription and capping in a dsRNA virus.
Elife, 4, 2015
|
|
3J9Q
 
 | | Atomic structures of a bactericidal contractile nanotube in its pre- and post-contraction states | | Descriptor: | sheath, tube | | Authors: | Ge, P, Scholl, D, Leiman, P.G, Yu, X, Miller, J.F, Zhou, Z.H. | | Deposit date: | 2015-02-17 | | Release date: | 2015-04-01 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Atomic structures of a bactericidal contractile nanotube in its pre- and postcontraction states.
Nat.Struct.Mol.Biol., 22, 2015
|
|
3JB5
 
 | |
3J9D
 
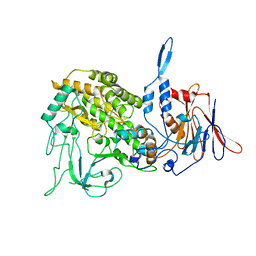 | | Atomic structure of a non-enveloped virus reveals pH sensors for a coordinated process of cell entry | | Descriptor: | Outer capsid protein VP2, ZINC ION | | Authors: | Zhang, X, Patel, A, Celma, C, Roy, P, Zhou, Z.H. | | Deposit date: | 2015-01-09 | | Release date: | 2015-12-09 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Atomic model of a nonenveloped virus reveals pH sensors for a coordinated process of cell entry.
Nat.Struct.Mol.Biol., 23, 2016
|
|
3JB3
 
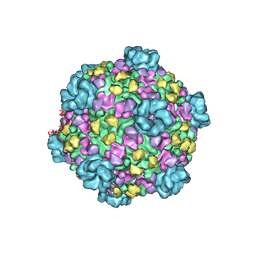 | | Atomic model of cytoplasmic polyhedrosis virus with SAM, GTP and ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Capsid protein VP1, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Yu, X.K, Jiang, J.S, Sun, J.C, Zhou, Z.H. | | Deposit date: | 2015-07-06 | | Release date: | 2015-08-12 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | A putative ATPase mediates RNA transcription and capping in a dsRNA virus.
Elife, 4, 2015
|
|
3J9C
 
 | | CryoEM single particle reconstruction of anthrax toxin protective antigen pore at 2.9 Angstrom resolution | | Descriptor: | CALCIUM ION, Protective antigen PA-63 | | Authors: | Jiang, J, Pentelute, B.L, Collier, R.J, Zhou, Z.H. | | Deposit date: | 2014-12-25 | | Release date: | 2015-03-11 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Atomic structure of anthrax protective antigen pore elucidates toxin translocation.
Nature, 521, 2015
|
|
3J9E
 
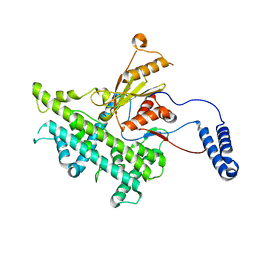 | | Atomic structure of a non-enveloped virus reveals pH sensors for a coordinated process of cell entry | | Descriptor: | VP5 | | Authors: | Zhang, X, Patel, A, Celma, C, Roy, P, Zhou, Z.H. | | Deposit date: | 2015-01-10 | | Release date: | 2015-12-09 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Atomic model of a nonenveloped virus reveals pH sensors for a coordinated process of cell entry.
Nat.Struct.Mol.Biol., 23, 2016
|
|
3J2P
 
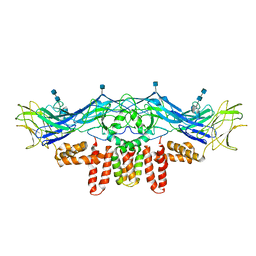 | | CryoEM structure of Dengue virus envelope protein heterotetramer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope protein E, ... | | Authors: | Zhang, X, Ge, P, Yu, X, Brannan, J.M, Bi, G, Zhang, Q, Schein, S, Zhou, Z.H. | | Deposit date: | 2012-11-30 | | Release date: | 2012-12-19 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structure of the mature dengue virus at 3.5-A resolution.
Nat.Struct.Mol.Biol., 20, 2012
|
|
8CXG
 
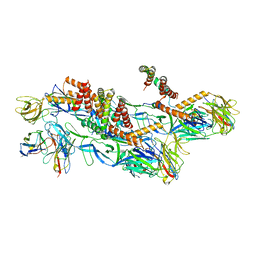 | | Structures of Zika Virus in Complex with Antibodies Targeting E Dimer Epitopes and Basis for Neutralization Efficacy | | Descriptor: | Ankyrin repeat family A protein 2,Envelope E protein, C8 scFv heavy chain, C8 scFv light chain, ... | | Authors: | Liu, W, Zhang, X.K, Gong, D.Y, Dai, X.H, Sharma, A, Zhang, T.H, Rey, F, Zhou, Z.H. | | Deposit date: | 2022-05-21 | | Release date: | 2023-06-07 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structures of Zika Virus in Complex with Antibodies Targeting E Dimer Epitopes and Basis for Neutralization Efficacy
To Be Published
|
|
8CXH
 
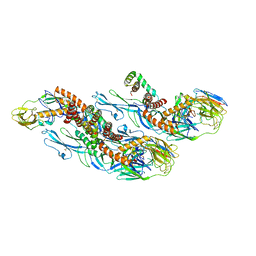 | | Structures of Zika Virus in Complex with Antibodies Targeting E Dimer Epitopes and Basis for Neutralization Efficacy | | Descriptor: | Ankyrin repeat family A protein 2,Envelope E protein, C10 heavy chain, C10 light chain, ... | | Authors: | Liu, W, Zhang, X.K, Gong, D.Y, Dai, X.H, Sharma, A, Zhang, T.H, Rey, F, Zhou, Z.H. | | Deposit date: | 2022-05-21 | | Release date: | 2023-06-07 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structures of Zika Virus in Complex with Antibodies Targeting E Dimer Epitopes and Basis for Neutralization Efficacy
To Be Published
|
|
8CXI
 
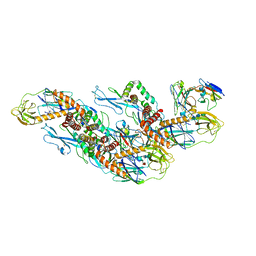 | | Structures of Zika Virus in Complex with Antibodies Targeting E Dimer Epitopes and Basis for Neutralization Efficacy | | Descriptor: | A11 heavy chain, A11 light chain, Ankyrin repeat family A protein 2,Envelope E protein, ... | | Authors: | Liu, W, Zhang, X.K, Gong, D.Y, Dai, X.H, Sharma, A, Zhang, T.H, Rey, F, Zhou, Z.H. | | Deposit date: | 2022-05-21 | | Release date: | 2023-06-07 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of Zika Virus in Complex with Antibodies Targeting E Dimer Epitopes and Basis for Neutralization Efficacy
To Be Published
|
|
8UD2
 
 | |
8UD3
 
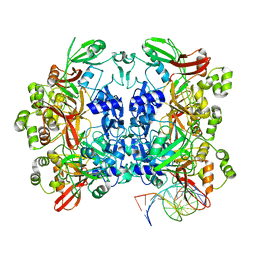 | | SARS-CoV-2 Nsp15 bound to poly(A/U) RNA, consensus form | | Descriptor: | Non-structural protein 15, RNA (35-MER) | | Authors: | Ito, F, Yang, H, Zhou, Z.H, Chen, X.S. | | Deposit date: | 2023-09-28 | | Release date: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.67 Å) | | Cite: | Structural basis for polyuridine tract recognition by SARS-CoV-2 Nsp15.
Protein Cell, 2024
|
|
8UD5
 
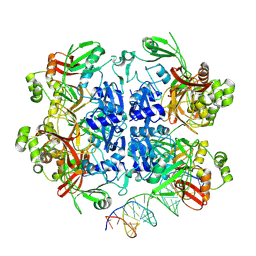 | | SARS-CoV-2 Nsp15 bound to poly(A/U) RNA, state 2 | | Descriptor: | Non-structural protein 15, RNA (35-MER) | | Authors: | Ito, F, Yang, H, Zhou, Z.H, Chen, X.S. | | Deposit date: | 2023-09-28 | | Release date: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Structural basis for polyuridine tract recognition by SARS-CoV-2 Nsp15.
Protein Cell, 2024
|
|
8UD4
 
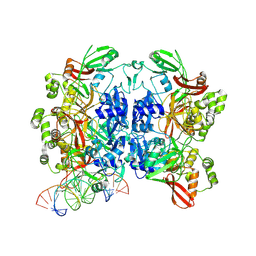 | | SARS-CoV-2 Nsp15 bound to poly(A/U) RNA, state 1 | | Descriptor: | Non-structural protein 15, RNA (35-MER) | | Authors: | Ito, F, Yang, H, Zhou, Z.H, Chen, X.S. | | Deposit date: | 2023-09-28 | | Release date: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Structural basis for polyuridine tract recognition by SARS-CoV-2 Nsp15.
Protein Cell, 2024
|
|
8F41
 
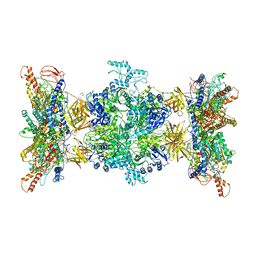 | | 3-methylcrotonyl-CoA carboxylase in filament, alpha-subunit centered | | Descriptor: | 3-methylcrotonyl-CoA carboxylase, alpha-subunit, beta-subunit, ... | | Authors: | Hu, J.J, Lee, J.K.J, Liu, Y.T, Yu, C, Huang, L, Afasizheva, I, Afasizhev, R, Zhou, Z.H. | | Deposit date: | 2022-11-10 | | Release date: | 2023-01-11 | | Last modified: | 2023-01-18 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Discovery, structure, and function of filamentous 3-methylcrotonyl-CoA carboxylase.
Structure, 31, 2023
|
|
8F3D
 
 | | 3-methylcrotonyl-CoA carboxylase in filament, beta-subunit centered | | Descriptor: | 3-methylcrotonyl-CoA carboxylase alpha-subunit, 3-methylcrotonyl-CoA carboxylase beta-subunit, 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL | | Authors: | Hu, J.J, Lee, J.K.J, Liu, Y.T, Yu, C, Huang, L, Afasizheva, I, Afasizhev, R, Zhou, Z.H. | | Deposit date: | 2022-11-09 | | Release date: | 2023-01-11 | | Last modified: | 2023-01-18 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Discovery, structure, and function of filamentous 3-methylcrotonyl-CoA carboxylase.
Structure, 31, 2023
|
|
6M5R
 
 | | The coordinates of the apo monomeric terminase complex | | Descriptor: | Tripartite terminase subunit 1, Tripartite terminase subunit 2, Tripartite terminase subunit 3, ... | | Authors: | Yang, Y.X, Yang, P, Wang, N, Chen, Z.H, Zhou, Z.H, Rao, Z.H, Wang, X.X. | | Deposit date: | 2020-03-11 | | Release date: | 2020-10-28 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Architecture of the herpesvirus genome-packaging complex and implications for DNA translocation.
Protein Cell, 11, 2020
|
|
6M5S
 
 | | The coordinates of the apo hexameric terminase complex | | Descriptor: | Tripartite terminase subunit 1, Tripartite terminase subunit 2, Tripartite terminase subunit 3, ... | | Authors: | Yang, Y.X, Yang, P, Wang, N, Chen, Z.H, Zhou, Z.H, Rao, Z.H, Wang, X.X. | | Deposit date: | 2020-03-11 | | Release date: | 2020-10-28 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Architecture of the herpesvirus genome-packaging complex and implications for DNA translocation.
Protein Cell, 11, 2020
|
|
6M5V
 
 | | The coordinate of the hexameric terminase complex in the presence of the ADP-BeF3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Yang, Y.X, Yang, P, Wang, N, Chen, Z.H, Zhou, Z.H, Rao, Z.H, Wang, X.X. | | Deposit date: | 2020-03-11 | | Release date: | 2020-10-28 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Architecture of the herpesvirus genome-packaging complex and implications for DNA translocation.
Protein Cell, 11, 2020
|
|
6M5T
 
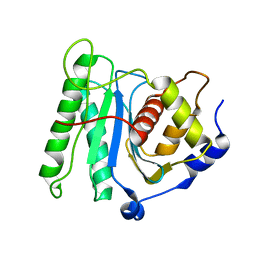 | | The coordinate of the nuclease domain of the apo terminase complex | | Descriptor: | Tripartite terminase subunit 3 | | Authors: | Yang, Y.X, Yang, P, Wang, N, Chen, Z.H, Zhou, Z.H, Rao, Z.H, Wang, X.X. | | Deposit date: | 2020-03-11 | | Release date: | 2020-10-28 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Architecture of the herpesvirus genome-packaging complex and implications for DNA translocation.
Protein Cell, 11, 2020
|
|
